Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
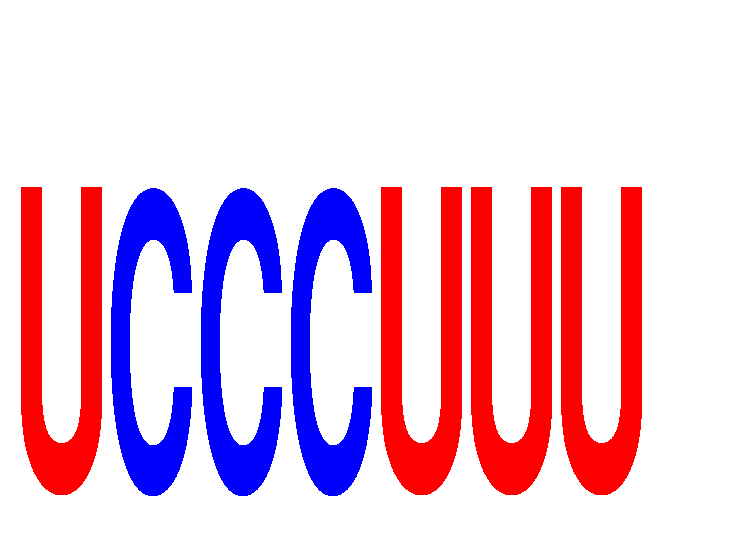
Results for UCCCUUU
Z-value: 0.19
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-204-5p
|
MIMAT0000237 |
|
mmu-miR-211-5p
|
MIMAT0000668 |
|
mmu-miR-7670-3p
|
MIMAT0029847 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_39490649 | 0.24 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr8_+_76899772 | 0.22 |
ENSMUST00000109913.2
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr7_-_131410495 | 0.22 |
ENSMUST00000121033.1
ENSMUST00000046306.8 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr13_+_75707484 | 0.20 |
ENSMUST00000001583.6
|
Ell2
|
elongation factor RNA polymerase II 2 |
| chr17_-_12769605 | 0.18 |
ENSMUST00000024599.7
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr9_-_72111827 | 0.16 |
ENSMUST00000183404.1
ENSMUST00000184783.1 |
Tcf12
|
transcription factor 12 |
| chrX_-_51018011 | 0.12 |
ENSMUST00000053593.7
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr10_-_9675163 | 0.11 |
ENSMUST00000100070.2
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr1_-_106714217 | 0.11 |
ENSMUST00000112751.1
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr2_-_103797617 | 0.10 |
ENSMUST00000028607.6
|
Caprin1
|
cell cycle associated protein 1 |
| chr15_-_78120011 | 0.10 |
ENSMUST00000019290.2
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr14_+_49066495 | 0.10 |
ENSMUST00000037473.4
|
Ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr9_-_104262900 | 0.10 |
ENSMUST00000035170.6
|
Dnajc13
|
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr13_+_41249841 | 0.10 |
ENSMUST00000165561.2
|
Smim13
|
small integral membrane protein 13 |
| chrX_-_56598069 | 0.10 |
ENSMUST00000059899.2
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr6_-_92214897 | 0.10 |
ENSMUST00000014694.8
|
Zfyve20
|
zinc finger, FYVE domain containing 20 |
| chr12_-_3309912 | 0.10 |
ENSMUST00000021001.8
|
Rab10
|
RAB10, member RAS oncogene family |
| chr4_+_13743424 | 0.10 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_-_34755229 | 0.10 |
ENSMUST00000102800.1
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr9_-_99140065 | 0.09 |
ENSMUST00000035037.7
|
Pik3cb
|
phosphatidylinositol 3-kinase, catalytic, beta polypeptide |
| chr1_+_64532790 | 0.09 |
ENSMUST00000049932.5
ENSMUST00000087366.4 ENSMUST00000171164.1 |
Creb1
|
cAMP responsive element binding protein 1 |
| chr9_-_75559604 | 0.09 |
ENSMUST00000072232.7
|
Tmod3
|
tropomodulin 3 |
| chr13_+_9093893 | 0.09 |
ENSMUST00000091829.2
|
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr18_+_51117754 | 0.09 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr18_-_64660981 | 0.09 |
ENSMUST00000025482.8
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr8_-_90348343 | 0.08 |
ENSMUST00000109621.3
|
Tox3
|
TOX high mobility group box family member 3 |
| chr13_+_31806627 | 0.08 |
ENSMUST00000062292.2
|
Foxc1
|
forkhead box C1 |
| chr10_-_117148474 | 0.07 |
ENSMUST00000020381.3
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr18_-_16809233 | 0.07 |
ENSMUST00000025166.7
|
Cdh2
|
cadherin 2 |
| chr5_-_123749371 | 0.07 |
ENSMUST00000182955.1
ENSMUST00000182489.1 ENSMUST00000050827.7 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr17_-_12507704 | 0.07 |
ENSMUST00000024595.2
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr6_+_141249161 | 0.07 |
ENSMUST00000043259.7
|
Pde3a
|
phosphodiesterase 3A, cGMP inhibited |
| chr12_-_34528844 | 0.07 |
ENSMUST00000110819.2
|
Hdac9
|
histone deacetylase 9 |
| chr1_-_170867761 | 0.07 |
ENSMUST00000027974.6
|
Atf6
|
activating transcription factor 6 |
| chr2_+_30061754 | 0.07 |
ENSMUST00000149578.1
ENSMUST00000102866.3 |
Set
|
SET nuclear oncogene |
| chr15_+_80711292 | 0.07 |
ENSMUST00000067689.7
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr5_+_124439891 | 0.07 |
ENSMUST00000059580.4
|
Setd8
|
SET domain containing (lysine methyltransferase) 8 |
| chr12_+_83763628 | 0.07 |
ENSMUST00000121733.1
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr8_-_123983120 | 0.06 |
ENSMUST00000075578.6
|
Abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr12_+_76370266 | 0.06 |
ENSMUST00000042779.3
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr13_-_38658991 | 0.06 |
ENSMUST00000001757.7
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr15_-_82912134 | 0.06 |
ENSMUST00000048966.5
ENSMUST00000109510.2 |
Tcf20
|
transcription factor 20 |
| chr7_+_141562162 | 0.06 |
ENSMUST00000003038.8
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr18_-_56975333 | 0.06 |
ENSMUST00000139243.2
ENSMUST00000025488.8 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr14_-_21052420 | 0.06 |
ENSMUST00000154460.1
|
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr6_-_118139094 | 0.06 |
ENSMUST00000049344.8
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr11_+_80209019 | 0.06 |
ENSMUST00000077451.7
ENSMUST00000055056.9 |
Rhot1
|
ras homolog gene family, member T1 |
| chr16_-_11203259 | 0.06 |
ENSMUST00000119953.1
|
Rsl1d1
|
ribosomal L1 domain containing 1 |
| chr3_-_51277470 | 0.06 |
ENSMUST00000108053.2
ENSMUST00000108051.1 |
Elf2
|
E74-like factor 2 |
| chr3_+_121953213 | 0.06 |
ENSMUST00000037958.7
ENSMUST00000128366.1 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr4_+_57637816 | 0.06 |
ENSMUST00000150412.1
|
Gm20459
|
predicted gene 20459 |
| chr1_-_43098622 | 0.06 |
ENSMUST00000095014.1
|
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr2_+_117121374 | 0.06 |
ENSMUST00000028829.6
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr19_-_12796108 | 0.05 |
ENSMUST00000038627.8
|
Zfp91
|
zinc finger protein 91 |
| chrX_+_159255782 | 0.05 |
ENSMUST00000126686.1
ENSMUST00000033671.6 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr12_-_3426700 | 0.05 |
ENSMUST00000180149.1
|
1110002L01Rik
|
RIKEN cDNA 1110002L01 gene |
| chr4_+_102760294 | 0.05 |
ENSMUST00000072481.5
ENSMUST00000156596.1 ENSMUST00000080728.6 ENSMUST00000106882.2 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr15_+_68928414 | 0.05 |
ENSMUST00000022954.6
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chrX_+_77511002 | 0.05 |
ENSMUST00000088217.5
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr13_+_74009419 | 0.05 |
ENSMUST00000022057.8
|
Tppp
|
tubulin polymerization promoting protein |
| chr12_-_84218835 | 0.05 |
ENSMUST00000046266.6
|
Elmsan1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr3_-_108911687 | 0.05 |
ENSMUST00000029480.8
|
Prpf38b
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
| chr1_-_55027473 | 0.05 |
ENSMUST00000027127.7
|
Sf3b1
|
splicing factor 3b, subunit 1 |
| chr16_+_57121705 | 0.05 |
ENSMUST00000166897.1
|
Tomm70a
|
translocase of outer mitochondrial membrane 70 homolog A (yeast) |
| chr6_-_51469869 | 0.05 |
ENSMUST00000114459.1
ENSMUST00000069949.6 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr13_+_37826225 | 0.04 |
ENSMUST00000128570.1
|
Rreb1
|
ras responsive element binding protein 1 |
| chr2_+_31572701 | 0.04 |
ENSMUST00000055244.6
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr1_-_79671966 | 0.04 |
ENSMUST00000162342.1
|
Ap1s3
|
adaptor-related protein complex AP-1, sigma 3 |
| chr2_+_84840612 | 0.04 |
ENSMUST00000111625.1
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr8_+_54954728 | 0.04 |
ENSMUST00000033915.7
|
Gpm6a
|
glycoprotein m6a |
| chr4_+_15265798 | 0.04 |
ENSMUST00000062684.8
|
Tmem64
|
transmembrane protein 64 |
| chr1_+_160906372 | 0.04 |
ENSMUST00000161609.1
|
Rc3h1
|
RING CCCH (C3H) domains 1 |
| chr4_+_106911470 | 0.04 |
ENSMUST00000030367.8
ENSMUST00000149926.1 |
Ssbp3
|
single-stranded DNA binding protein 3 |
| chr13_-_17805093 | 0.04 |
ENSMUST00000042365.7
|
Cdk13
|
cyclin-dependent kinase 13 |
| chr2_+_3713449 | 0.04 |
ENSMUST00000027965.4
|
Fam107b
|
family with sequence similarity 107, member B |
| chr2_-_130840091 | 0.04 |
ENSMUST00000044766.8
ENSMUST00000138990.1 ENSMUST00000120316.1 ENSMUST00000110243.1 |
4930402H24Rik
|
RIKEN cDNA 4930402H24 gene |
| chr14_+_47663756 | 0.04 |
ENSMUST00000022391.7
|
Ktn1
|
kinectin 1 |
| chr9_+_104002546 | 0.04 |
ENSMUST00000035167.8
ENSMUST00000117054.1 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr4_+_57845240 | 0.04 |
ENSMUST00000102903.1
ENSMUST00000107598.3 |
Akap2
|
A kinase (PRKA) anchor protein 2 |
| chr9_-_60511003 | 0.04 |
ENSMUST00000098660.3
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr2_-_52424826 | 0.04 |
ENSMUST00000036541.7
|
Arl5a
|
ADP-ribosylation factor-like 5A |
| chr1_-_84284548 | 0.04 |
ENSMUST00000177458.1
ENSMUST00000168574.2 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr2_+_109917639 | 0.04 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr8_+_106059562 | 0.04 |
ENSMUST00000109308.1
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr2_+_15055274 | 0.04 |
ENSMUST00000069870.3
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr5_+_96373955 | 0.04 |
ENSMUST00000036019.4
|
Fras1
|
Fraser syndrome 1 homolog (human) |
| chr17_-_6782775 | 0.03 |
ENSMUST00000064234.6
|
Ezr
|
ezrin |
| chr11_-_76846968 | 0.03 |
ENSMUST00000021201.5
|
Cpd
|
carboxypeptidase D |
| chr8_+_109868586 | 0.03 |
ENSMUST00000179721.1
ENSMUST00000034175.4 |
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr5_+_76974683 | 0.03 |
ENSMUST00000101087.3
ENSMUST00000120550.1 |
Srp72
|
signal recognition particle 72 |
| chr3_-_89913144 | 0.03 |
ENSMUST00000029559.6
|
Il6ra
|
interleukin 6 receptor, alpha |
| chr3_-_95217690 | 0.03 |
ENSMUST00000107209.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr4_+_41135743 | 0.03 |
ENSMUST00000040008.3
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr11_+_54438188 | 0.03 |
ENSMUST00000046835.7
|
Fnip1
|
folliculin interacting protein 1 |
| chr13_-_75943812 | 0.03 |
ENSMUST00000022078.5
ENSMUST00000109606.1 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr16_-_26105777 | 0.03 |
ENSMUST00000039990.5
|
Leprel1
|
leprecan-like 1 |
| chr8_-_87804411 | 0.03 |
ENSMUST00000165770.2
|
Zfp423
|
zinc finger protein 423 |
| chrX_-_23365044 | 0.03 |
ENSMUST00000115313.1
|
Klhl13
|
kelch-like 13 |
| chr15_+_25622525 | 0.03 |
ENSMUST00000110457.1
ENSMUST00000137601.1 |
Myo10
|
myosin X |
| chr5_+_110176640 | 0.03 |
ENSMUST00000112512.1
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr2_-_5714490 | 0.03 |
ENSMUST00000044009.7
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr3_+_60501252 | 0.03 |
ENSMUST00000099087.2
|
Mbnl1
|
muscleblind-like 1 (Drosophila) |
| chr6_-_30304513 | 0.03 |
ENSMUST00000094543.2
ENSMUST00000102993.3 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr10_-_29535857 | 0.03 |
ENSMUST00000092623.3
|
Rspo3
|
R-spondin 3 homolog (Xenopus laevis) |
| chr10_-_75517324 | 0.03 |
ENSMUST00000039796.7
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr19_+_18749983 | 0.03 |
ENSMUST00000040489.7
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr1_+_127774164 | 0.03 |
ENSMUST00000027587.8
ENSMUST00000112570.1 |
Ccnt2
|
cyclin T2 |
| chr7_+_132859225 | 0.03 |
ENSMUST00000084497.5
ENSMUST00000181577.1 ENSMUST00000106161.1 |
Fam175b
|
family with sequence similarity 175, member B |
| chr16_-_23988852 | 0.02 |
ENSMUST00000023151.5
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr10_+_128092771 | 0.02 |
ENSMUST00000170054.2
ENSMUST00000045621.8 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr9_-_113708209 | 0.02 |
ENSMUST00000111861.3
ENSMUST00000035086.6 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr6_-_138043411 | 0.02 |
ENSMUST00000111873.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chrX_-_64276937 | 0.02 |
ENSMUST00000114679.1
ENSMUST00000069926.7 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr13_+_45507438 | 0.02 |
ENSMUST00000000260.6
|
Gmpr
|
guanosine monophosphate reductase |
| chr5_-_62766153 | 0.02 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_+_152081529 | 0.02 |
ENSMUST00000064061.3
|
Scrt2
|
scratch homolog 2, zinc finger protein (Drosophila) |
| chr4_+_28813125 | 0.02 |
ENSMUST00000029964.5
ENSMUST00000080934.4 |
Epha7
|
Eph receptor A7 |
| chr3_-_138489372 | 0.02 |
ENSMUST00000029804.8
|
Metap1
|
methionyl aminopeptidase 1 |
| chr15_+_80173642 | 0.02 |
ENSMUST00000044970.6
|
Mgat3
|
mannoside acetylglucosaminyltransferase 3 |
| chr6_+_41605482 | 0.02 |
ENSMUST00000114732.2
|
Ephb6
|
Eph receptor B6 |
| chr14_-_100149764 | 0.02 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr19_+_47067721 | 0.02 |
ENSMUST00000026027.5
|
Taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_-_92943485 | 0.02 |
ENSMUST00000113438.1
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 |
| chr2_-_37647199 | 0.02 |
ENSMUST00000028279.3
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr16_+_8830093 | 0.02 |
ENSMUST00000023150.5
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr10_+_128805652 | 0.02 |
ENSMUST00000026410.1
|
Dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr10_-_120476469 | 0.02 |
ENSMUST00000072777.7
ENSMUST00000159699.1 |
Hmga2
|
high mobility group AT-hook 2 |
| chr5_+_105699945 | 0.02 |
ENSMUST00000060531.9
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr3_-_56183678 | 0.02 |
ENSMUST00000029374.6
|
Nbea
|
neurobeachin |
| chrX_+_93654863 | 0.01 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr15_-_98607611 | 0.01 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr5_+_108461222 | 0.01 |
ENSMUST00000046975.5
ENSMUST00000112597.1 |
Pcgf3
|
polycomb group ring finger 3 |
| chr2_-_69586021 | 0.01 |
ENSMUST00000100051.2
ENSMUST00000092551.4 ENSMUST00000080953.5 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr18_-_46280820 | 0.01 |
ENSMUST00000025354.3
|
Pggt1b
|
protein geranylgeranyltransferase type I, beta subunit |
| chr16_+_17451981 | 0.01 |
ENSMUST00000006293.3
|
Crkl
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like |
| chr12_+_53248677 | 0.01 |
ENSMUST00000101432.2
|
Npas3
|
neuronal PAS domain protein 3 |
| chr18_+_23752333 | 0.01 |
ENSMUST00000170802.1
ENSMUST00000155708.1 ENSMUST00000118826.2 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_156992021 | 0.01 |
ENSMUST00000109558.1
ENSMUST00000069600.6 ENSMUST00000072298.6 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr18_+_76944384 | 0.01 |
ENSMUST00000156454.1
ENSMUST00000150990.1 ENSMUST00000026485.7 ENSMUST00000148955.1 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr2_+_25180737 | 0.01 |
ENSMUST00000104999.2
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr2_-_57114970 | 0.01 |
ENSMUST00000028166.2
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_+_51415986 | 0.01 |
ENSMUST00000029303.7
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr2_-_73892530 | 0.01 |
ENSMUST00000136958.1
ENSMUST00000112010.2 ENSMUST00000128531.1 ENSMUST00000112017.1 |
Atf2
|
activating transcription factor 2 |
| chr9_+_44604844 | 0.01 |
ENSMUST00000170489.1
|
Ddx6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 |
| chr11_+_20201406 | 0.01 |
ENSMUST00000020358.5
ENSMUST00000109602.1 ENSMUST00000109601.1 |
Rab1
|
RAB1, member RAS oncogene family |
| chr15_+_6708372 | 0.01 |
ENSMUST00000061656.6
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chrX_+_163908982 | 0.01 |
ENSMUST00000069041.8
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_+_99535439 | 0.01 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chrX_-_144688180 | 0.01 |
ENSMUST00000040184.3
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr5_-_51553896 | 0.01 |
ENSMUST00000132734.1
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr10_-_67285180 | 0.01 |
ENSMUST00000159002.1
ENSMUST00000077839.6 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr17_-_80480435 | 0.01 |
ENSMUST00000068714.5
|
Sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr2_-_164911586 | 0.01 |
ENSMUST00000041361.7
|
Zfp335
|
zinc finger protein 335 |
| chr6_+_108783059 | 0.01 |
ENSMUST00000032196.6
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr11_-_69560186 | 0.01 |
ENSMUST00000004036.5
|
Efnb3
|
ephrin B3 |
| chr17_-_32284088 | 0.01 |
ENSMUST00000119123.1
ENSMUST00000003726.8 ENSMUST00000121285.1 ENSMUST00000120276.1 ENSMUST00000114475.2 |
Brd4
|
bromodomain containing 4 |
| chr16_+_41532999 | 0.01 |
ENSMUST00000099761.3
|
Lsamp
|
limbic system-associated membrane protein |
| chr1_+_187997821 | 0.01 |
ENSMUST00000027906.6
|
Esrrg
|
estrogen-related receptor gamma |
| chr11_+_17159263 | 0.01 |
ENSMUST00000102880.4
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
| chr18_-_88894322 | 0.01 |
ENSMUST00000070116.5
ENSMUST00000125362.1 |
Socs6
|
suppressor of cytokine signaling 6 |
| chr14_-_25769033 | 0.01 |
ENSMUST00000069180.7
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr4_-_143212691 | 0.01 |
ENSMUST00000105778.1
ENSMUST00000134791.1 |
Prdm2
|
PR domain containing 2, with ZNF domain |
| chr13_-_28953690 | 0.01 |
ENSMUST00000067230.5
|
Sox4
|
SRY-box containing gene 4 |
| chr1_-_131279544 | 0.01 |
ENSMUST00000062108.3
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr5_+_115341225 | 0.01 |
ENSMUST00000031508.4
|
Triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr6_-_31563978 | 0.00 |
ENSMUST00000026698.7
|
Podxl
|
podocalyxin-like |
| chr4_+_152338619 | 0.00 |
ENSMUST00000030775.5
ENSMUST00000164662.1 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr14_+_11553523 | 0.00 |
ENSMUST00000022264.6
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr9_-_22052021 | 0.00 |
ENSMUST00000003501.7
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) |
| chr8_-_26119125 | 0.00 |
ENSMUST00000037182.7
|
Hook3
|
hook homolog 3 (Drosophila) |
| chr2_+_107290590 | 0.00 |
ENSMUST00000037012.2
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr8_-_121652895 | 0.00 |
ENSMUST00000046386.4
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr19_-_42612766 | 0.00 |
ENSMUST00000166128.1
ENSMUST00000026190.7 ENSMUST00000164786.1 |
Loxl4
|
lysyl oxidase-like 4 |
| chr3_+_28263205 | 0.00 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr7_+_57590503 | 0.00 |
ENSMUST00000085240.4
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr18_+_9212856 | 0.00 |
ENSMUST00000041080.5
|
Fzd8
|
frizzled homolog 8 (Drosophila) |
| chr10_-_123076367 | 0.00 |
ENSMUST00000073792.3
ENSMUST00000170935.1 ENSMUST00000037557.7 |
Mon2
|
MON2 homolog (yeast) |
| chr5_+_114774677 | 0.00 |
ENSMUST00000102578.4
|
Ankrd13a
|
ankyrin repeat domain 13a |
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.0 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.1 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) regulation of cellular pH reduction(GO:0032847) |
| 0.0 | 0.1 | GO:1990869 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.0 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |