Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
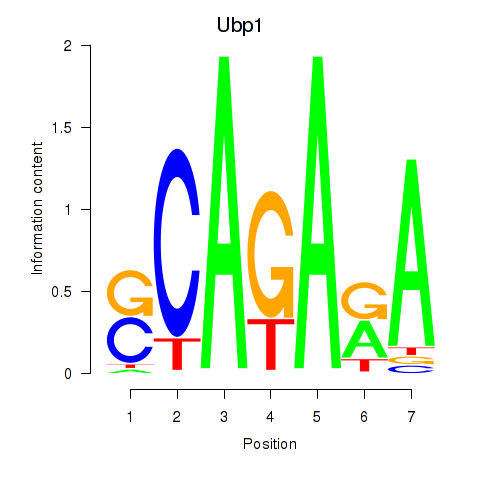
Results for Ubp1
Z-value: 1.18
Transcription factors associated with Ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ubp1
|
ENSMUSG00000009741.8 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ubp1 | mm10_v2_chr9_+_113930934_113930987 | -0.66 | 9.9e-06 | Click! |
Activity profile of Ubp1 motif
Sorted Z-values of Ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13623967 | 11.39 |
ENSMUST00000108525.2
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr4_-_115496129 | 7.53 |
ENSMUST00000030487.2
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr19_-_7802578 | 5.87 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr7_-_14123042 | 5.02 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr5_+_130448801 | 4.70 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr7_+_26808880 | 4.24 |
ENSMUST00000040944.7
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr10_+_127759780 | 4.23 |
ENSMUST00000128247.1
|
RP23-386P10.11
|
Protein Rdh9 |
| chr6_-_55175019 | 4.14 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr1_-_150465563 | 4.11 |
ENSMUST00000164600.1
ENSMUST00000111902.2 ENSMUST00000111901.2 ENSMUST00000006171.9 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr5_-_87092546 | 4.03 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr11_+_101367542 | 3.99 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr19_+_39007019 | 3.87 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr13_-_55426783 | 3.76 |
ENSMUST00000021948.8
|
F12
|
coagulation factor XII (Hageman factor) |
| chr19_-_7966000 | 3.72 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr19_+_3986564 | 3.62 |
ENSMUST00000054030.7
|
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr6_-_48572660 | 3.61 |
ENSMUST00000009425.4
|
Rarres2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr19_-_8218832 | 3.48 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr1_-_169747634 | 3.39 |
ENSMUST00000027991.5
ENSMUST00000111357.1 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr19_-_40073731 | 3.38 |
ENSMUST00000048959.3
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr4_-_96664112 | 3.38 |
ENSMUST00000030299.7
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr3_+_130617645 | 3.36 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr3_+_130617448 | 3.36 |
ENSMUST00000166187.1
ENSMUST00000072271.6 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr9_+_46268601 | 3.25 |
ENSMUST00000121598.1
|
Apoa5
|
apolipoprotein A-V |
| chr13_-_19307551 | 3.24 |
ENSMUST00000103561.1
|
Tcrg-C2
|
T-cell receptor gamma, constant 2 |
| chr7_+_27119909 | 3.18 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr7_+_107567445 | 3.18 |
ENSMUST00000120990.1
|
Olfml1
|
olfactomedin-like 1 |
| chr8_-_4105764 | 3.15 |
ENSMUST00000138439.1
ENSMUST00000145007.1 |
Cd209f
|
CD209f antigen |
| chrX_+_139800795 | 3.15 |
ENSMUST00000054889.3
|
Cldn2
|
claudin 2 |
| chr13_+_4436094 | 3.05 |
ENSMUST00000156277.1
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr7_+_26061495 | 2.99 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr6_+_68161415 | 2.95 |
ENSMUST00000168090.1
|
Igkv1-115
|
immunoglobulin kappa variable 1-115 |
| chr10_+_127898515 | 2.82 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr5_-_86926521 | 2.80 |
ENSMUST00000031183.2
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr7_+_68275970 | 2.80 |
ENSMUST00000153805.1
|
Fam169b
|
family with sequence similarity 169, member B |
| chr7_-_13989588 | 2.66 |
ENSMUST00000165167.1
ENSMUST00000108520.2 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr3_+_138277489 | 2.61 |
ENSMUST00000004232.9
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr11_-_84167466 | 2.56 |
ENSMUST00000050771.7
|
Gm11437
|
predicted gene 11437 |
| chr13_-_22219820 | 2.54 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr15_+_76696725 | 2.52 |
ENSMUST00000023203.4
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr9_+_119102463 | 2.47 |
ENSMUST00000140326.1
ENSMUST00000165231.1 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr7_-_12998172 | 2.44 |
ENSMUST00000120903.1
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr5_-_87424201 | 2.44 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr8_-_93197799 | 2.43 |
ENSMUST00000034172.7
|
Ces1d
|
carboxylesterase 1D |
| chr13_-_55426769 | 2.34 |
ENSMUST00000170921.1
|
F12
|
coagulation factor XII (Hageman factor) |
| chr19_+_4711153 | 2.30 |
ENSMUST00000008991.6
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr10_+_127849917 | 2.29 |
ENSMUST00000077530.2
|
Rdh19
|
retinol dehydrogenase 19 |
| chr3_-_98893209 | 2.28 |
ENSMUST00000029464.7
|
Hao2
|
hydroxyacid oxidase 2 |
| chr7_-_12998140 | 2.27 |
ENSMUST00000032539.7
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr14_+_53649387 | 2.25 |
ENSMUST00000103660.2
|
Trav15-2-dv6-2
|
T cell receptor alpha variable 15-2-DV6-2 |
| chr7_-_19692596 | 2.24 |
ENSMUST00000108451.2
ENSMUST00000045035.4 |
Apoc1
|
apolipoprotein C-I |
| chr11_-_110095937 | 2.23 |
ENSMUST00000106664.3
ENSMUST00000046223.7 ENSMUST00000106662.1 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr3_+_129836729 | 2.16 |
ENSMUST00000077918.5
|
Cfi
|
complement component factor i |
| chr14_+_65970804 | 2.12 |
ENSMUST00000138191.1
|
Clu
|
clusterin |
| chr4_-_140617062 | 2.07 |
ENSMUST00000154979.1
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr8_+_4134733 | 2.06 |
ENSMUST00000130372.1
|
Cd209g
|
CD209g antigen |
| chr16_-_33056174 | 2.06 |
ENSMUST00000115100.1
ENSMUST00000040309.8 |
Iqcg
|
IQ motif containing G |
| chr10_-_89533550 | 2.06 |
ENSMUST00000105297.1
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_-_105600103 | 2.06 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr4_-_6275629 | 2.04 |
ENSMUST00000029905.1
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr19_-_8405060 | 2.02 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr13_-_93637961 | 2.02 |
ENSMUST00000099309.4
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr1_+_130865669 | 1.95 |
ENSMUST00000038829.5
|
Faim3
|
Fas apoptotic inhibitory molecule 3 |
| chrX_-_103821940 | 1.91 |
ENSMUST00000042664.5
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2 |
| chr13_-_41847626 | 1.90 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_+_24736673 | 1.89 |
ENSMUST00000101800.5
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr7_+_26835305 | 1.88 |
ENSMUST00000005685.8
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr1_+_88166004 | 1.86 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr8_-_3467617 | 1.80 |
ENSMUST00000111081.3
ENSMUST00000118194.1 ENSMUST00000004686.6 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr18_+_33464163 | 1.79 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr3_-_107986408 | 1.78 |
ENSMUST00000012348.2
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chr7_-_114562945 | 1.76 |
ENSMUST00000119712.1
ENSMUST00000032908.8 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr11_+_99041237 | 1.74 |
ENSMUST00000017637.6
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr5_-_25100624 | 1.73 |
ENSMUST00000030784.7
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr3_+_19957037 | 1.72 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr9_-_121916288 | 1.71 |
ENSMUST00000062474.4
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr1_+_171214013 | 1.68 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr14_-_65953728 | 1.67 |
ENSMUST00000042046.3
|
Scara3
|
scavenger receptor class A, member 3 |
| chr11_-_75422586 | 1.67 |
ENSMUST00000138661.1
ENSMUST00000000769.7 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr1_+_164796723 | 1.66 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr14_-_7994563 | 1.65 |
ENSMUST00000026315.7
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr1_-_162866502 | 1.63 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_-_148038270 | 1.62 |
ENSMUST00000132070.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr3_+_146597077 | 1.60 |
ENSMUST00000029837.7
ENSMUST00000121133.1 |
Uox
|
urate oxidase |
| chr14_+_51007911 | 1.59 |
ENSMUST00000022424.6
|
Rnase10
|
ribonuclease, RNase A family, 10 (non-active) |
| chr6_+_41538218 | 1.59 |
ENSMUST00000103291.1
|
Trbc1
|
T cell receptor beta, constant region 1 |
| chr11_+_97030130 | 1.58 |
ENSMUST00000153482.1
|
Scrn2
|
secernin 2 |
| chr3_+_19957088 | 1.58 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr6_+_41546730 | 1.56 |
ENSMUST00000103299.1
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr17_+_24736639 | 1.56 |
ENSMUST00000115262.1
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr17_+_79626669 | 1.54 |
ENSMUST00000086570.1
|
4921513D11Rik
|
RIKEN cDNA 4921513D11 gene |
| chr8_-_45358737 | 1.53 |
ENSMUST00000155230.1
ENSMUST00000135912.1 |
Fam149a
|
family with sequence similarity 149, member A |
| chr5_+_7960445 | 1.52 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chr8_+_70083509 | 1.49 |
ENSMUST00000007738.9
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr14_+_41151442 | 1.49 |
ENSMUST00000047095.2
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chrX_+_73483602 | 1.46 |
ENSMUST00000033741.8
ENSMUST00000169489.1 |
Bgn
|
biglycan |
| chrX_-_85776606 | 1.45 |
ENSMUST00000142152.1
ENSMUST00000156390.1 ENSMUST00000113978.2 |
Gyk
|
glycerol kinase |
| chr5_-_151369172 | 1.45 |
ENSMUST00000067770.3
|
D730045B01Rik
|
RIKEN cDNA D730045B01 gene |
| chr6_-_85933379 | 1.45 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr2_-_32424005 | 1.43 |
ENSMUST00000113307.2
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr6_-_113434757 | 1.43 |
ENSMUST00000113091.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr14_-_56234650 | 1.42 |
ENSMUST00000015585.2
|
Gzmc
|
granzyme C |
| chr10_+_127759721 | 1.41 |
ENSMUST00000073639.5
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr10_-_95415283 | 1.40 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr8_+_13026024 | 1.40 |
ENSMUST00000033820.3
|
F7
|
coagulation factor VII |
| chr11_-_73326807 | 1.39 |
ENSMUST00000134079.1
|
Aspa
|
aspartoacylase |
| chr3_+_118562129 | 1.38 |
ENSMUST00000039177.7
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr1_+_88070765 | 1.38 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr5_-_38502107 | 1.37 |
ENSMUST00000005238.6
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr14_-_70520254 | 1.37 |
ENSMUST00000022693.7
|
Bmp1
|
bone morphogenetic protein 1 |
| chr19_-_11266122 | 1.36 |
ENSMUST00000169159.1
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr14_-_10453443 | 1.36 |
ENSMUST00000161302.1
|
Fhit
|
fragile histidine triad gene |
| chr2_-_25501717 | 1.34 |
ENSMUST00000015227.3
|
C8g
|
complement component 8, gamma polypeptide |
| chr17_-_46487641 | 1.34 |
ENSMUST00000047034.8
|
Ttbk1
|
tau tubulin kinase 1 |
| chr19_-_24861828 | 1.33 |
ENSMUST00000047666.4
|
Pgm5
|
phosphoglucomutase 5 |
| chr11_-_115062177 | 1.33 |
ENSMUST00000062787.7
|
Cd300e
|
CD300e antigen |
| chr16_+_17894197 | 1.33 |
ENSMUST00000046937.2
|
Tssk1
|
testis-specific serine kinase 1 |
| chrX_+_59999436 | 1.32 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr8_-_120634379 | 1.32 |
ENSMUST00000123927.1
|
1190005I06Rik
|
RIKEN cDNA 1190005I06 gene |
| chr7_+_130936172 | 1.32 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr11_-_110095974 | 1.31 |
ENSMUST00000100287.2
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr1_+_171213969 | 1.31 |
ENSMUST00000005820.4
ENSMUST00000075469.5 ENSMUST00000155126.1 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_-_36989504 | 1.31 |
ENSMUST00000169189.1
|
H2-M5
|
histocompatibility 2, M region locus 5 |
| chr2_-_35100677 | 1.30 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr18_-_74961252 | 1.29 |
ENSMUST00000066532.4
|
Lipg
|
lipase, endothelial |
| chr11_-_70812539 | 1.28 |
ENSMUST00000074572.6
ENSMUST00000108534.2 |
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr9_-_103222063 | 1.28 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr15_+_10714836 | 1.28 |
ENSMUST00000180604.1
|
4930556M19Rik
|
RIKEN cDNA 4930556M19 gene |
| chr2_-_128967725 | 1.27 |
ENSMUST00000099385.2
|
Gm10762
|
predicted gene 10762 |
| chr13_+_91741507 | 1.27 |
ENSMUST00000022120.4
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr15_+_31572179 | 1.26 |
ENSMUST00000161088.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr3_+_94362444 | 1.26 |
ENSMUST00000169433.1
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr7_+_4337710 | 1.24 |
ENSMUST00000006792.4
|
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr5_+_90518932 | 1.23 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr9_-_103219823 | 1.23 |
ENSMUST00000168142.1
|
Trf
|
transferrin |
| chr7_-_140102326 | 1.22 |
ENSMUST00000128527.1
|
Fuom
|
fucose mutarotase |
| chr10_+_62071014 | 1.22 |
ENSMUST00000053865.5
|
Gm5424
|
predicted gene 5424 |
| chr10_-_127070254 | 1.22 |
ENSMUST00000060991.4
|
Tspan31
|
tetraspanin 31 |
| chr14_-_70429072 | 1.22 |
ENSMUST00000048129.4
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr11_-_77519186 | 1.22 |
ENSMUST00000100807.2
|
Gm10392
|
predicted gene 10392 |
| chr7_+_140835018 | 1.21 |
ENSMUST00000106050.1
ENSMUST00000026554.4 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr1_+_181051232 | 1.21 |
ENSMUST00000036819.6
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr14_+_53743104 | 1.20 |
ENSMUST00000103667.4
|
Trav16
|
T cell receptor alpha variable 16 |
| chr11_+_51651179 | 1.18 |
ENSMUST00000170689.1
|
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr16_+_37580137 | 1.18 |
ENSMUST00000160847.1
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr1_-_172895048 | 1.18 |
ENSMUST00000027824.5
|
Apcs
|
serum amyloid P-component |
| chr2_+_147364989 | 1.17 |
ENSMUST00000109968.2
|
Pax1
|
paired box gene 1 |
| chr10_+_106470281 | 1.16 |
ENSMUST00000029404.9
ENSMUST00000169303.1 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_+_65311257 | 1.16 |
ENSMUST00000027083.6
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr5_+_30814722 | 1.16 |
ENSMUST00000114724.1
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr19_+_12599953 | 1.15 |
ENSMUST00000181868.1
ENSMUST00000092931.6 |
Gm4952
|
predicted gene 4952 |
| chr3_+_29082539 | 1.14 |
ENSMUST00000119598.1
ENSMUST00000118531.1 |
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr5_+_30588078 | 1.14 |
ENSMUST00000066295.2
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr9_-_106685892 | 1.13 |
ENSMUST00000169068.1
ENSMUST00000046735.4 |
Tex264
|
testis expressed gene 264 |
| chr8_+_94472763 | 1.12 |
ENSMUST00000053085.5
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr17_-_36168532 | 1.11 |
ENSMUST00000040467.8
|
Gm8909
|
predicted gene 8909 |
| chr11_-_75422524 | 1.11 |
ENSMUST00000125982.1
ENSMUST00000137103.1 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_69695521 | 1.11 |
ENSMUST00000181261.1
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr16_-_46120238 | 1.11 |
ENSMUST00000023336.9
|
Cd96
|
CD96 antigen |
| chr19_+_44989073 | 1.10 |
ENSMUST00000026225.8
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr3_+_19957240 | 1.10 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr11_-_118342500 | 1.10 |
ENSMUST00000103024.3
|
BC100451
|
cDNA sequence BC100451 |
| chr11_+_115440540 | 1.09 |
ENSMUST00000093914.4
|
4933422H20Rik
|
RIKEN cDNA 4933422H20 gene |
| chr6_-_124888192 | 1.08 |
ENSMUST00000024044.6
|
Cd4
|
CD4 antigen |
| chr16_-_21787796 | 1.08 |
ENSMUST00000023559.5
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
| chr12_-_103989917 | 1.07 |
ENSMUST00000151709.2
ENSMUST00000176246.1 ENSMUST00000074693.7 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr5_+_33104219 | 1.07 |
ENSMUST00000011178.2
|
Slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr6_-_90224438 | 1.06 |
ENSMUST00000076086.2
|
Vmn1r53
|
vomeronasal 1 receptor 53 |
| chr7_+_64287665 | 1.06 |
ENSMUST00000032736.4
|
Mtmr10
|
myotubularin related protein 10 |
| chr11_-_50931612 | 1.05 |
ENSMUST00000109124.3
|
Zfp354b
|
zinc finger protein 354B |
| chr14_+_12189943 | 1.04 |
ENSMUST00000119888.1
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr14_-_30923547 | 1.04 |
ENSMUST00000170415.1
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr6_+_15720654 | 1.04 |
ENSMUST00000101663.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr2_-_24049389 | 1.04 |
ENSMUST00000051416.5
|
Hnmt
|
histamine N-methyltransferase |
| chr11_-_73326472 | 1.04 |
ENSMUST00000155630.2
|
Aspa
|
aspartoacylase |
| chr13_-_47043116 | 1.03 |
ENSMUST00000110118.1
ENSMUST00000124948.1 ENSMUST00000021806.3 ENSMUST00000136864.1 |
Tpmt
|
thiopurine methyltransferase |
| chr17_+_31386218 | 1.03 |
ENSMUST00000047168.5
|
Pde9a
|
phosphodiesterase 9A |
| chr4_-_108071327 | 1.03 |
ENSMUST00000106701.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr11_+_97029925 | 1.01 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr6_-_42373254 | 1.01 |
ENSMUST00000073387.2
|
Epha1
|
Eph receptor A1 |
| chr12_-_28623282 | 1.01 |
ENSMUST00000036136.7
|
Colec11
|
collectin sub-family member 11 |
| chr12_-_103242143 | 1.01 |
ENSMUST00000074416.3
|
Prima1
|
proline rich membrane anchor 1 |
| chr11_-_101171302 | 1.00 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr11_+_82045705 | 1.00 |
ENSMUST00000021011.2
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr6_-_35326123 | 1.00 |
ENSMUST00000051176.7
|
Fam180a
|
family with sequence similarity 180, member A |
| chr11_-_12464850 | 0.98 |
ENSMUST00000109650.1
|
Cobl
|
cordon-bleu WH2 repeat |
| chr16_+_93683184 | 0.98 |
ENSMUST00000039620.6
|
Cbr3
|
carbonyl reductase 3 |
| chr17_-_47437365 | 0.98 |
ENSMUST00000061885.8
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr4_-_129227883 | 0.97 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr6_-_114969986 | 0.97 |
ENSMUST00000139640.1
|
Vgll4
|
vestigial like 4 (Drosophila) |
| chr12_-_31559969 | 0.96 |
ENSMUST00000001253.7
|
Slc26a4
|
solute carrier family 26, member 4 |
| chr18_+_37473538 | 0.96 |
ENSMUST00000050034.1
|
Pcdhb15
|
protocadherin beta 15 |
| chr10_+_23894688 | 0.96 |
ENSMUST00000041416.7
|
Vnn1
|
vanin 1 |
| chr2_+_25428699 | 0.95 |
ENSMUST00000102919.3
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_-_12464804 | 0.94 |
ENSMUST00000172919.1
|
Cobl
|
cordon-bleu WH2 repeat |
| chr11_+_70700473 | 0.94 |
ENSMUST00000152618.2
ENSMUST00000102554.1 ENSMUST00000094499.4 ENSMUST00000072187.5 |
Kif1c
|
kinesin family member 1C |
| chr16_+_11313812 | 0.94 |
ENSMUST00000023140.5
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr13_-_34077992 | 0.94 |
ENSMUST00000056427.8
|
Tubb2a
|
tubulin, beta 2A class IIA |
| chr6_+_48589560 | 0.93 |
ENSMUST00000181661.1
|
Gm5111
|
predicted gene 5111 |
| chr2_+_163472545 | 0.93 |
ENSMUST00000065731.4
|
2310001K24Rik
|
RIKEN cDNA 2310001K24 gene |
| chr11_+_83703991 | 0.92 |
ENSMUST00000092836.5
|
Wfdc17
|
WAP four-disulfide core domain 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0002541 | plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.2 | 4.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.1 | 3.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 1.1 | 15.1 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 3.0 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.0 | 4.0 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.9 | 2.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.9 | 2.8 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.9 | 3.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.9 | 3.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.8 | 3.2 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.7 | 2.1 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.7 | 4.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.7 | 2.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.6 | 7.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.5 | 4.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.5 | 1.4 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 1.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) thymidine metabolic process(GO:0046104) |
| 0.5 | 2.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 2.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 2.0 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 1.2 | GO:0000239 | pachytene(GO:0000239) |
| 0.4 | 2.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 1.6 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.4 | 1.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.4 | 1.8 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.4 | 2.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 2.7 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 3.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 2.0 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.3 | 1.0 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 9.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 1.3 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 0.9 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.3 | 0.9 | GO:0070347 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.3 | 1.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 3.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 2.0 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 1.1 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.3 | 1.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 3.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.3 | 0.8 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 2.8 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 1.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 0.7 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 1.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 0.7 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 0.9 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.2 | 1.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.2 | 1.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 2.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 0.4 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 0.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.6 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 0.8 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.2 | 1.5 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 8.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.7 | GO:0097494 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of vesicle size(GO:0097494) |
| 0.2 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 1.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.2 | 0.5 | GO:0051795 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.2 | 2.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 0.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.2 | 0.3 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 1.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.2 | 0.6 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.9 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 0.8 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.4 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.6 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.0 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 2.3 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.8 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 1.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 0.7 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.5 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 1.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.8 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.0 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 2.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 1.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.6 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 1.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.7 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 0.4 | GO:0071727 | response to diacyl bacterial lipopeptide(GO:0071724) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to diacyl bacterial lipopeptide(GO:0071726) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 3.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.3 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.6 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 1.8 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.4 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.4 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.4 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.1 | 0.3 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.1 | 0.3 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.8 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.6 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.1 | 0.8 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.9 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.4 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.8 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 2.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.3 | GO:2000387 | regulation of type IV hypersensitivity(GO:0001807) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) |
| 0.1 | 0.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.5 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.5 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.8 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 2.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 2.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.4 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.0 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.8 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 1.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.7 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.7 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 0.3 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 2.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.5 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 0.3 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.1 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 1.3 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.1 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.2 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 2.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.0 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.2 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.7 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.3 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.6 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.2 | GO:1903896 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.3 | GO:0089700 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) protein kinase D signaling(GO:0089700) |
| 0.0 | 1.4 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 1.2 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.2 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 2.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.6 | GO:0043470 | regulation of glycolytic process(GO:0006110) regulation of carbohydrate catabolic process(GO:0043470) regulation of cellular carbohydrate catabolic process(GO:0043471) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 2.2 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 1.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.0 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 1.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.5 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.6 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.1 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:1904415 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.7 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.2 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.0 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.3 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.8 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 3.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.7 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.0 | GO:0052572 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) response to host(GO:0075136) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.4 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 1.9 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:1904305 | negative regulation of gastro-intestinal system smooth muscle contraction(GO:1904305) negative regulation of small intestine smooth muscle contraction(GO:1904348) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.3 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0060159 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 2.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 0.9 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 1.3 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.3 | 1.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 2.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 1.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 0.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 3.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 2.1 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 5.5 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 1.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.5 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 4.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.8 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 6.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 6.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 2.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 3.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 6.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 5.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 5.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.2 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 32.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 4.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 3.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 13.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 19.1 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 2.2 | 6.7 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.8 | 7.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.3 | 4.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.2 | 7.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.1 | 16.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.0 | 3.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.9 | 5.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.9 | 2.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.9 | 6.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.8 | 2.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.6 | 3.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.6 | 2.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.5 | 2.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.5 | 5.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 3.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 4.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 12.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 1.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.4 | 1.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.4 | 1.6 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.4 | 4.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 2.5 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 6.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 13.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 1.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 1.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 2.3 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 1.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 1.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 2.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 0.8 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.3 | 0.8 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 1.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 5.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 1.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.3 | 4.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.2 | 1.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.7 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.2 | 1.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.7 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 5.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 1.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 1.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 0.8 | GO:0042903 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.2 | 1.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.4 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.2 | 0.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 1.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 0.7 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 0.7 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 4.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 3.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.3 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.1 | 0.6 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.7 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 2.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 2.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.9 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 1.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 1.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.4 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 2.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 2.4 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) S100 protein binding(GO:0044548) |
| 0.1 | 1.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 10.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 2.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.4 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 1.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 12.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 3.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 3.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 1.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 5.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 4.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 5.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 3.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |