Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
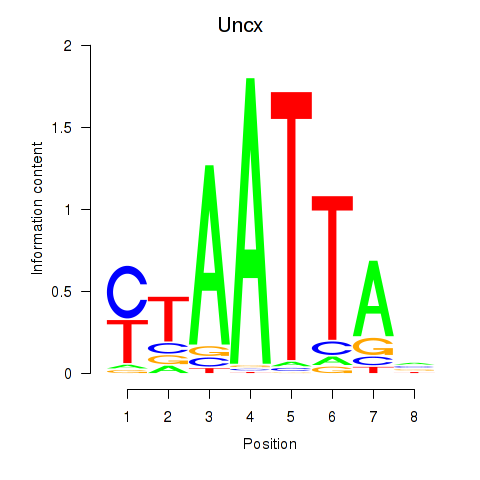
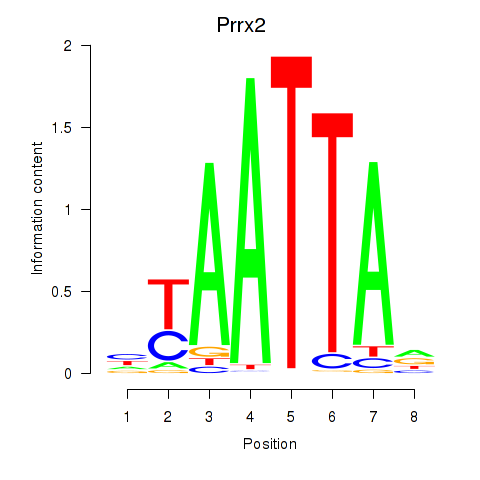
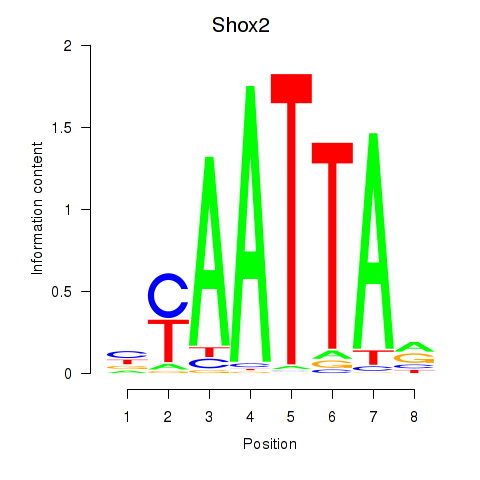
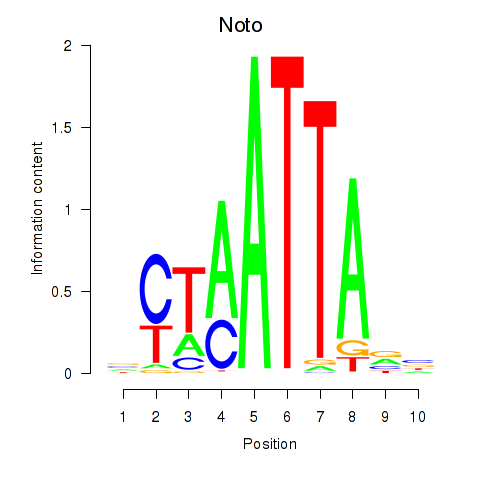
Results for Vsx1_Uncx_Prrx2_Shox2_Noto
Z-value: 0.83
Transcription factors associated with Vsx1_Uncx_Prrx2_Shox2_Noto
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vsx1
|
ENSMUSG00000033080.8 | visual system homeobox 1 |
|
Uncx
|
ENSMUSG00000029546.11 | UNC homeobox |
|
Prrx2
|
ENSMUSG00000039476.7 | paired related homeobox 2 |
|
Shox2
|
ENSMUSG00000027833.10 | short stature homeobox 2 |
|
Noto
|
ENSMUSG00000068302.7 | notochord homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prrx2 | mm10_v2_chr2_+_30834972_30834972 | 0.38 | 2.2e-02 | Click! |
| Shox2 | mm10_v2_chr3_-_66981279_66981318 | 0.25 | 1.3e-01 | Click! |
| Uncx | mm10_v2_chr5_+_139543889_139543904 | -0.10 | 5.5e-01 | Click! |
| Noto | mm10_v2_chr6_+_85423886_85423886 | 0.09 | 6.2e-01 | Click! |
| Vsx1 | mm10_v2_chr2_-_150689360_150689360 | -0.07 | 6.9e-01 | Click! |
Activity profile of Vsx1_Uncx_Prrx2_Shox2_Noto motif
Sorted Z-values of Vsx1_Uncx_Prrx2_Shox2_Noto motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_34356421 | 5.87 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr14_+_27000362 | 5.85 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_-_168767029 | 5.48 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr2_-_168767136 | 5.21 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr15_-_79285502 | 4.80 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr3_-_14778452 | 4.11 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr15_-_79285470 | 3.86 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr12_+_109545390 | 3.52 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr16_-_42340595 | 3.10 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr14_-_118052235 | 2.94 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr2_-_72813665 | 2.55 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr11_-_87359011 | 2.30 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr2_+_25372315 | 2.28 |
ENSMUST00000028329.6
ENSMUST00000114293.2 ENSMUST00000100323.2 |
Sapcd2
|
suppressor APC domain containing 2 |
| chr11_+_67200052 | 2.21 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr1_+_172555932 | 2.16 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr5_+_34999070 | 2.14 |
ENSMUST00000114280.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_+_34999111 | 2.11 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chrX_+_133850980 | 2.07 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr5_+_34999046 | 2.03 |
ENSMUST00000114281.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr9_+_96258697 | 1.91 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr6_-_145865302 | 1.83 |
ENSMUST00000111703.1
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr10_-_45470201 | 1.83 |
ENSMUST00000079390.6
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr3_-_116253467 | 1.80 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr6_-_30958990 | 1.73 |
ENSMUST00000101589.3
|
Klf14
|
Kruppel-like factor 14 |
| chr17_+_34592248 | 1.68 |
ENSMUST00000038149.6
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr15_-_66812593 | 1.66 |
ENSMUST00000100572.3
|
Sla
|
src-like adaptor |
| chrX_-_74246534 | 1.57 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chrX_+_150547375 | 1.53 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr1_-_132390301 | 1.42 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr7_+_144838590 | 1.39 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chr6_+_6248659 | 1.39 |
ENSMUST00000181633.1
ENSMUST00000176283.1 ENSMUST00000175814.1 ENSMUST00000181192.1 |
Gm20619
|
predicted gene 20619 |
| chr11_+_67200137 | 1.35 |
ENSMUST00000129018.1
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr17_+_46161021 | 1.33 |
ENSMUST00000024748.7
ENSMUST00000172170.1 |
Gtpbp2
|
GTP binding protein 2 |
| chr4_-_24430838 | 1.30 |
ENSMUST00000183964.1
|
RP23-35K5.2
|
RP23-35K5.2 |
| chrX_-_7978027 | 1.30 |
ENSMUST00000125418.1
|
Gata1
|
GATA binding protein 1 |
| chr13_+_23575753 | 1.25 |
ENSMUST00000105105.1
|
Hist1h3d
|
histone cluster 1, H3d |
| chrX_-_139871637 | 1.24 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chr8_-_120228221 | 1.23 |
ENSMUST00000183235.1
|
A330074K22Rik
|
RIKEN cDNA A330074K22 gene |
| chrX_+_56454871 | 1.19 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr2_+_152754156 | 1.18 |
ENSMUST00000010020.5
|
Cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr3_-_100489324 | 1.17 |
ENSMUST00000061455.8
|
Fam46c
|
family with sequence similarity 46, member C |
| chr16_-_74411292 | 1.09 |
ENSMUST00000117200.1
|
Robo2
|
roundabout homolog 2 (Drosophila) |
| chr6_-_102464667 | 1.08 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr18_+_44104407 | 1.05 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr2_+_84734050 | 1.03 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr1_-_126830632 | 1.02 |
ENSMUST00000112583.1
ENSMUST00000094609.3 |
Nckap5
|
NCK-associated protein 5 |
| chr1_-_78968079 | 1.01 |
ENSMUST00000049117.5
|
Gm5830
|
predicted pseudogene 5830 |
| chr2_+_80638798 | 1.01 |
ENSMUST00000028382.6
ENSMUST00000124377.1 |
Nup35
|
nucleoporin 35 |
| chr1_+_40515362 | 0.99 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr7_-_100855403 | 0.99 |
ENSMUST00000156855.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr14_+_80000292 | 0.98 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chrX_+_9885622 | 0.97 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr17_-_31198958 | 0.96 |
ENSMUST00000114549.2
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr5_+_90490714 | 0.96 |
ENSMUST00000042755.3
|
Afp
|
alpha fetoprotein |
| chr8_-_3624989 | 0.96 |
ENSMUST00000142431.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_+_27165233 | 0.95 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr8_+_121116163 | 0.93 |
ENSMUST00000054691.6
|
Foxc2
|
forkhead box C2 |
| chr2_+_86007778 | 0.93 |
ENSMUST00000062166.1
|
Olfr1032
|
olfactory receptor 1032 |
| chr6_-_138079916 | 0.91 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr19_-_55241236 | 0.90 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr8_-_3625274 | 0.87 |
ENSMUST00000004749.6
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr8_+_83666827 | 0.86 |
ENSMUST00000019608.5
|
Ptger1
|
prostaglandin E receptor 1 (subtype EP1) |
| chr4_+_34893772 | 0.86 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr3_+_122044428 | 0.85 |
ENSMUST00000013995.8
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chrX_+_159303266 | 0.85 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chrX_-_150814265 | 0.83 |
ENSMUST00000026302.6
ENSMUST00000129768.1 ENSMUST00000112699.2 |
Maged2
|
melanoma antigen, family D, 2 |
| chr4_-_99654983 | 0.82 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr3_-_130730375 | 0.82 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr2_-_164171113 | 0.78 |
ENSMUST00000045196.3
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr3_+_55782500 | 0.77 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr7_-_115824699 | 0.77 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr10_-_24092320 | 0.77 |
ENSMUST00000092654.2
|
Taar8b
|
trace amine-associated receptor 8B |
| chr6_+_134929118 | 0.77 |
ENSMUST00000185152.1
ENSMUST00000184504.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr13_+_8202885 | 0.75 |
ENSMUST00000139438.1
ENSMUST00000135574.1 |
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr8_-_62123106 | 0.75 |
ENSMUST00000034052.6
ENSMUST00000034054.7 |
Anxa10
|
annexin A10 |
| chr17_+_7025837 | 0.75 |
ENSMUST00000089120.5
|
Gm1604b
|
predicted gene 1604b |
| chr17_+_12119274 | 0.74 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr19_-_15924560 | 0.74 |
ENSMUST00000162053.1
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr3_-_50443603 | 0.73 |
ENSMUST00000029297.4
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr3_-_72967854 | 0.72 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr4_-_154636831 | 0.71 |
ENSMUST00000030902.6
ENSMUST00000105637.1 ENSMUST00000070313.7 ENSMUST00000105636.1 ENSMUST00000105638.2 ENSMUST00000097759.2 ENSMUST00000124771.1 |
Prdm16
|
PR domain containing 16 |
| chr11_+_102604370 | 0.71 |
ENSMUST00000057893.5
|
Fzd2
|
frizzled homolog 2 (Drosophila) |
| chr8_-_31918203 | 0.71 |
ENSMUST00000073884.4
|
Nrg1
|
neuregulin 1 |
| chr17_-_24073479 | 0.70 |
ENSMUST00000017090.5
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr6_+_125552948 | 0.69 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr3_+_90600203 | 0.68 |
ENSMUST00000001047.7
|
S100a3
|
S100 calcium binding protein A3 |
| chr12_+_102128718 | 0.68 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr2_-_170194033 | 0.67 |
ENSMUST00000180625.1
|
Gm17619
|
predicted gene, 17619 |
| chr17_-_45733843 | 0.67 |
ENSMUST00000178179.1
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr12_-_11208948 | 0.67 |
ENSMUST00000049877.1
|
Msgn1
|
mesogenin 1 |
| chr9_-_53975246 | 0.66 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr4_+_13751297 | 0.66 |
ENSMUST00000105566.2
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_-_72986716 | 0.65 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr5_-_148392810 | 0.65 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr3_+_108092789 | 0.64 |
ENSMUST00000058669.8
ENSMUST00000145101.1 |
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr7_-_45830776 | 0.62 |
ENSMUST00000107723.2
ENSMUST00000131384.1 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr10_-_80421847 | 0.60 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr11_+_67171027 | 0.59 |
ENSMUST00000170159.1
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr9_+_119063429 | 0.58 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr11_+_67171095 | 0.58 |
ENSMUST00000018641.7
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr3_+_108093109 | 0.58 |
ENSMUST00000151326.1
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr18_+_77332394 | 0.56 |
ENSMUST00000148341.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr10_+_88091070 | 0.55 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr5_+_13398688 | 0.55 |
ENSMUST00000125629.1
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_159839729 | 0.54 |
ENSMUST00000068952.5
|
Wls
|
wntless homolog (Drosophila) |
| chr4_+_19818722 | 0.54 |
ENSMUST00000035890.7
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr11_+_103133333 | 0.54 |
ENSMUST00000124928.1
ENSMUST00000062530.4 |
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr7_-_37773555 | 0.54 |
ENSMUST00000176534.1
|
Zfp536
|
zinc finger protein 536 |
| chr5_-_138170992 | 0.54 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr19_-_5560570 | 0.53 |
ENSMUST00000025861.1
|
Ovol1
|
OVO homolog-like 1 (Drosophila) |
| chr15_+_92597104 | 0.53 |
ENSMUST00000035399.8
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr18_+_24603952 | 0.52 |
ENSMUST00000025120.6
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr10_+_127420867 | 0.52 |
ENSMUST00000064793.6
|
R3hdm2
|
R3H domain containing 2 |
| chr14_-_110755100 | 0.52 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr4_-_4138432 | 0.51 |
ENSMUST00000070375.7
|
Penk
|
preproenkephalin |
| chr5_-_138171248 | 0.51 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr8_+_107031218 | 0.51 |
ENSMUST00000034388.9
|
Vps4a
|
vacuolar protein sorting 4a (yeast) |
| chr6_+_123262107 | 0.51 |
ENSMUST00000032240.2
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr13_-_97747373 | 0.50 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr2_+_4017727 | 0.50 |
ENSMUST00000177457.1
|
Frmd4a
|
FERM domain containing 4A |
| chr13_+_118714678 | 0.50 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chr17_+_71019503 | 0.50 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr5_-_138171216 | 0.49 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr3_+_125404072 | 0.49 |
ENSMUST00000173932.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr3_-_17230976 | 0.49 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr1_-_126830786 | 0.49 |
ENSMUST00000162646.1
|
Nckap5
|
NCK-associated protein 5 |
| chr2_+_130012336 | 0.49 |
ENSMUST00000110299.2
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr3_-_126998408 | 0.49 |
ENSMUST00000182764.1
ENSMUST00000044443.8 |
Ank2
|
ankyrin 2, brain |
| chrX_+_21484532 | 0.49 |
ENSMUST00000089188.2
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr9_+_24283433 | 0.48 |
ENSMUST00000154644.1
|
Npsr1
|
neuropeptide S receptor 1 |
| chr17_+_71019548 | 0.48 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr17_-_29888570 | 0.48 |
ENSMUST00000171691.1
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr3_-_88410295 | 0.47 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chr13_-_97747399 | 0.47 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr8_-_24725762 | 0.46 |
ENSMUST00000171438.1
ENSMUST00000171611.1 ENSMUST00000033958.7 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr17_-_48432723 | 0.45 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr8_+_4238733 | 0.45 |
ENSMUST00000110998.2
ENSMUST00000062686.4 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr11_+_103133303 | 0.44 |
ENSMUST00000107037.1
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr7_-_46667375 | 0.44 |
ENSMUST00000107669.2
|
Tph1
|
tryptophan hydroxylase 1 |
| chr13_-_113663670 | 0.44 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr9_+_55541148 | 0.44 |
ENSMUST00000034869.4
|
Isl2
|
insulin related protein 2 (islet 2) |
| chr17_+_49615104 | 0.44 |
ENSMUST00000162854.1
|
Kif6
|
kinesin family member 6 |
| chr13_-_114458720 | 0.43 |
ENSMUST00000022287.5
|
Fst
|
follistatin |
| chr18_+_54990280 | 0.43 |
ENSMUST00000181538.1
|
Gm4221
|
predicted gene 4221 |
| chr5_-_66514815 | 0.42 |
ENSMUST00000161879.1
ENSMUST00000159357.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr17_+_17402672 | 0.42 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr3_+_134236483 | 0.41 |
ENSMUST00000181904.1
ENSMUST00000053048.9 |
Cxxc4
|
CXXC finger 4 |
| chr18_-_24603464 | 0.41 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr1_-_172027269 | 0.41 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr5_+_19907502 | 0.40 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr18_-_24603791 | 0.39 |
ENSMUST00000070726.3
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr1_-_150164943 | 0.39 |
ENSMUST00000181308.1
|
Gm26687
|
predicted gene, 26687 |
| chr11_+_76904475 | 0.39 |
ENSMUST00000142166.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_+_85979312 | 0.39 |
ENSMUST00000170610.1
|
Olfr1030
|
olfactory receptor 1030 |
| chr4_-_14621805 | 0.38 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr11_+_67277124 | 0.38 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr10_+_128337761 | 0.38 |
ENSMUST00000005826.7
|
Cs
|
citrate synthase |
| chr18_-_54990124 | 0.38 |
ENSMUST00000064763.5
|
Zfp608
|
zinc finger protein 608 |
| chr2_+_125068118 | 0.36 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr10_-_92375367 | 0.36 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr12_+_38780284 | 0.36 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chr4_-_149126688 | 0.35 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr7_-_5014645 | 0.35 |
ENSMUST00000165320.1
|
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr11_-_74724670 | 0.35 |
ENSMUST00000021091.8
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr12_-_40037387 | 0.35 |
ENSMUST00000146905.1
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr9_+_65890237 | 0.34 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr11_+_52098681 | 0.34 |
ENSMUST00000020608.2
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
| chr6_+_92816460 | 0.34 |
ENSMUST00000057977.3
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr15_-_102366314 | 0.34 |
ENSMUST00000078508.5
|
Sp7
|
Sp7 transcription factor 7 |
| chrX_-_143933204 | 0.33 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr5_-_103629279 | 0.33 |
ENSMUST00000031263.1
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr3_+_125404292 | 0.33 |
ENSMUST00000144344.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr17_+_35533194 | 0.33 |
ENSMUST00000025273.8
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr11_-_97700327 | 0.33 |
ENSMUST00000018681.7
|
Pcgf2
|
polycomb group ring finger 2 |
| chr4_-_14621494 | 0.32 |
ENSMUST00000149633.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr8_-_5105232 | 0.32 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr15_-_100424092 | 0.32 |
ENSMUST00000154676.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chrX_-_102157065 | 0.32 |
ENSMUST00000056904.2
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr11_-_98053415 | 0.32 |
ENSMUST00000017544.2
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr5_+_14025305 | 0.32 |
ENSMUST00000073957.6
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr1_-_66945361 | 0.31 |
ENSMUST00000160100.1
|
Myl1
|
myosin, light polypeptide 1 |
| chrM_+_2743 | 0.31 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr13_+_44121167 | 0.31 |
ENSMUST00000163056.1
ENSMUST00000159595.1 |
Gm5083
|
predicted gene 5083 |
| chr13_-_102906046 | 0.31 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_+_154960915 | 0.31 |
ENSMUST00000049621.6
|
Hes5
|
hairy and enhancer of split 5 (Drosophila) |
| chrX_+_159708593 | 0.31 |
ENSMUST00000080394.6
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr15_+_21111452 | 0.31 |
ENSMUST00000075132.6
|
Cdh12
|
cadherin 12 |
| chr15_-_8710409 | 0.30 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chrX_-_134111852 | 0.30 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr2_+_153938218 | 0.30 |
ENSMUST00000109757.1
|
Bpifb4
|
BPI fold containing family B, member 4 |
| chrX_-_150589844 | 0.29 |
ENSMUST00000112725.1
ENSMUST00000112720.1 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr2_+_57997884 | 0.29 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr5_-_138187177 | 0.29 |
ENSMUST00000110937.1
ENSMUST00000139276.1 ENSMUST00000048698.7 ENSMUST00000123415.1 |
Taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_-_163725123 | 0.28 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr15_+_18818895 | 0.28 |
ENSMUST00000166873.2
|
Cdh10
|
cadherin 10 |
| chr3_-_155055341 | 0.28 |
ENSMUST00000064076.3
|
Tnni3k
|
TNNI3 interacting kinase |
| chr11_-_53470479 | 0.28 |
ENSMUST00000057722.2
|
Gm9837
|
predicted gene 9837 |
| chr2_+_105130883 | 0.28 |
ENSMUST00000111098.1
ENSMUST00000111099.1 |
Wt1
|
Wilms tumor 1 homolog |
| chr14_+_53324632 | 0.28 |
ENSMUST00000178100.1
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr18_+_23415400 | 0.27 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx1_Uncx_Prrx2_Shox2_Noto
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 1.0 | 2.9 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.8 | 7.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.6 | 1.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.6 | 10.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.5 | 2.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.5 | 4.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 1.3 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.4 | 2.4 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.4 | 8.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 1.0 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.3 | 0.9 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.7 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 0.6 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 1.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.6 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.2 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.5 | GO:1903774 | mitotic cytokinesis checkpoint(GO:0044878) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 1.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.8 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.5 | GO:2000293 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 3.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.5 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.5 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.5 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.7 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.6 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.3 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.9 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 4.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.4 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 1.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.7 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.7 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 1.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.7 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0021764 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 1.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) olfactory nerve development(GO:0021553) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.0 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 1.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 2.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 1.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.5 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.0 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 1.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.0 | GO:0048880 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 1.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 1.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.2 | 5.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 6.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 11.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 3.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 2.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0071820 | N-box binding(GO:0071820) |
| 0.5 | 4.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 1.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.4 | 3.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 1.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 1.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 0.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 2.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 6.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 1.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 1.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 6.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 5.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 11.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 4.4 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 9.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |