Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
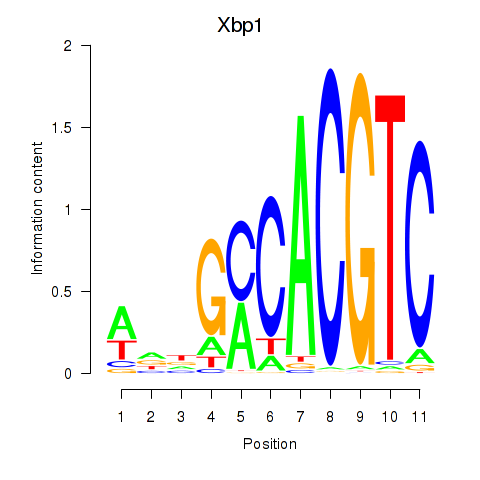
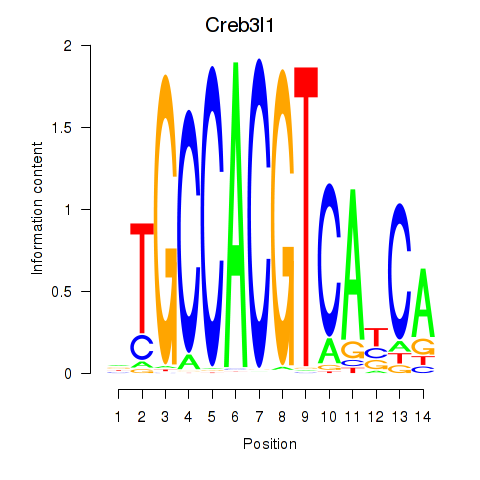
Results for Xbp1_Creb3l1
Z-value: 0.59
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Xbp1
|
ENSMUSG00000020484.12 | X-box binding protein 1 |
|
Creb3l1
|
ENSMUSG00000027230.9 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | mm10_v2_chr2_-_92024502_92024529 | 0.53 | 9.3e-04 | Click! |
| Xbp1 | mm10_v2_chr11_+_5520652_5520659 | 0.09 | 6.0e-01 | Click! |
Activity profile of Xbp1_Creb3l1 motif
Sorted Z-values of Xbp1_Creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_128400448 | 1.87 |
ENSMUST00000167859.1
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_142578093 | 1.80 |
ENSMUST00000149974.1
ENSMUST00000152754.1 |
H19
|
H19 fetal liver mRNA |
| chr14_-_60086832 | 1.80 |
ENSMUST00000080368.5
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr7_-_142578139 | 1.74 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr15_+_79516396 | 1.61 |
ENSMUST00000010974.7
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr7_-_45092130 | 1.45 |
ENSMUST00000148175.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_-_45092198 | 1.34 |
ENSMUST00000140449.1
ENSMUST00000117546.1 ENSMUST00000019683.3 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr17_-_26199008 | 1.15 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr10_-_84533884 | 1.00 |
ENSMUST00000053871.3
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr7_-_45211877 | 1.00 |
ENSMUST00000033057.7
|
Dkkl1
|
dickkopf-like 1 |
| chr10_+_128908907 | 0.98 |
ENSMUST00000105229.1
|
Cd63
|
CD63 antigen |
| chr7_-_45091713 | 0.91 |
ENSMUST00000141576.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr10_-_84533968 | 0.90 |
ENSMUST00000167671.1
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr12_-_36042476 | 0.86 |
ENSMUST00000020896.8
|
Tspan13
|
tetraspanin 13 |
| chr3_+_93520473 | 0.80 |
ENSMUST00000029515.4
|
S100a11
|
S100 calcium binding protein A11 (calgizzarin) |
| chr4_-_155653184 | 0.71 |
ENSMUST00000030937.1
|
Mmp23
|
matrix metallopeptidase 23 |
| chr9_+_72925622 | 0.70 |
ENSMUST00000038489.5
|
Pygo1
|
pygopus 1 |
| chr3_-_27710413 | 0.70 |
ENSMUST00000046157.4
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr18_+_57354733 | 0.66 |
ENSMUST00000025490.8
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr18_-_52529692 | 0.63 |
ENSMUST00000025409.7
|
Lox
|
lysyl oxidase |
| chr14_-_79301623 | 0.56 |
ENSMUST00000022595.7
|
Rgcc
|
regulator of cell cycle |
| chr2_+_150786735 | 0.52 |
ENSMUST00000045441.7
|
Pygb
|
brain glycogen phosphorylase |
| chr6_-_72958465 | 0.51 |
ENSMUST00000114050.1
|
Tmsb10
|
thymosin, beta 10 |
| chr11_+_69095217 | 0.51 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr2_-_105399286 | 0.49 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr4_-_108406676 | 0.45 |
ENSMUST00000184609.1
|
Gpx7
|
glutathione peroxidase 7 |
| chr14_+_20674311 | 0.44 |
ENSMUST00000048657.8
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr18_-_52529847 | 0.42 |
ENSMUST00000171470.1
|
Lox
|
lysyl oxidase |
| chr6_-_101377342 | 0.42 |
ENSMUST00000151175.1
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr6_-_52204415 | 0.41 |
ENSMUST00000048794.6
|
Hoxa5
|
homeobox A5 |
| chr18_+_65581704 | 0.37 |
ENSMUST00000182979.1
|
Zfp532
|
zinc finger protein 532 |
| chr9_+_57504012 | 0.37 |
ENSMUST00000080514.7
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr8_+_106168857 | 0.36 |
ENSMUST00000034378.3
|
Slc7a6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 |
| chr10_+_91082940 | 0.36 |
ENSMUST00000020150.3
|
Ikbip
|
IKBKB interacting protein |
| chr6_-_29212240 | 0.36 |
ENSMUST00000160878.1
ENSMUST00000078155.5 |
Impdh1
|
inosine 5'-phosphate dehydrogenase 1 |
| chr15_-_32244632 | 0.35 |
ENSMUST00000181536.1
|
0610007N19Rik
|
RIKEN cDNA 0610007N19 |
| chr16_-_56886131 | 0.34 |
ENSMUST00000023435.5
|
Tmem45a
|
transmembrane protein 45a |
| chr5_-_35679416 | 0.33 |
ENSMUST00000114233.2
|
Htra3
|
HtrA serine peptidase 3 |
| chr4_-_129640959 | 0.32 |
ENSMUST00000132217.1
ENSMUST00000130017.1 ENSMUST00000154105.1 |
Txlna
|
taxilin alpha |
| chr12_+_88953399 | 0.32 |
ENSMUST00000057634.7
|
Nrxn3
|
neurexin III |
| chr5_+_137288273 | 0.32 |
ENSMUST00000024099.4
ENSMUST00000085934.3 |
Ache
|
acetylcholinesterase |
| chr10_-_91082653 | 0.32 |
ENSMUST00000159110.1
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr10_-_91082704 | 0.30 |
ENSMUST00000162618.1
ENSMUST00000020157.6 ENSMUST00000160788.1 |
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr5_+_144190284 | 0.29 |
ENSMUST00000060747.7
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr7_+_27258725 | 0.28 |
ENSMUST00000079258.6
|
Numbl
|
numb-like |
| chr4_-_129640691 | 0.28 |
ENSMUST00000084264.5
|
Txlna
|
taxilin alpha |
| chr4_-_11386679 | 0.27 |
ENSMUST00000043781.7
ENSMUST00000108310.1 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr13_-_38528412 | 0.26 |
ENSMUST00000035988.8
|
Txndc5
|
thioredoxin domain containing 5 |
| chr4_-_129641060 | 0.26 |
ENSMUST00000046425.9
ENSMUST00000133803.1 |
Txlna
|
taxilin alpha |
| chr3_-_19628669 | 0.26 |
ENSMUST00000119133.1
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr3_+_96697076 | 0.25 |
ENSMUST00000162778.2
ENSMUST00000064900.9 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr4_-_11386757 | 0.25 |
ENSMUST00000108313.1
ENSMUST00000108311.2 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr8_+_70594466 | 0.25 |
ENSMUST00000019283.9
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr11_-_69948145 | 0.25 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr10_-_85102487 | 0.25 |
ENSMUST00000059383.6
|
Fhl4
|
four and a half LIM domains 4 |
| chr10_-_128211788 | 0.24 |
ENSMUST00000061995.8
|
Spryd4
|
SPRY domain containing 4 |
| chr8_+_125995102 | 0.24 |
ENSMUST00000046765.8
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr3_-_97767916 | 0.24 |
ENSMUST00000045243.8
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr11_-_108343917 | 0.24 |
ENSMUST00000059595.4
|
Prkca
|
protein kinase C, alpha |
| chr7_+_28693032 | 0.23 |
ENSMUST00000151227.1
ENSMUST00000108281.1 |
Fbxo27
|
F-box protein 27 |
| chr5_-_135251209 | 0.23 |
ENSMUST00000062572.2
|
Fzd9
|
frizzled homolog 9 (Drosophila) |
| chr19_+_5088534 | 0.23 |
ENSMUST00000025811.4
|
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_-_55226768 | 0.23 |
ENSMUST00000027121.8
ENSMUST00000114428.2 |
Rftn2
|
raftlin family member 2 |
| chr6_-_126740151 | 0.23 |
ENSMUST00000112242.1
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chrX_+_56454871 | 0.22 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr7_+_35555367 | 0.22 |
ENSMUST00000181932.1
|
B230322F03Rik
|
RIKEN cDNA B230322F03 gene |
| chr7_+_82867327 | 0.22 |
ENSMUST00000082237.5
|
Mex3b
|
mex3 homolog B (C. elegans) |
| chr1_-_163313661 | 0.22 |
ENSMUST00000174397.1
ENSMUST00000075805.6 ENSMUST00000027878.7 |
Prrx1
|
paired related homeobox 1 |
| chr6_-_72958097 | 0.21 |
ENSMUST00000114049.1
|
Tmsb10
|
thymosin, beta 10 |
| chr7_-_44869788 | 0.21 |
ENSMUST00000046575.9
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr11_-_100970887 | 0.21 |
ENSMUST00000060792.5
|
Ptrf
|
polymerase I and transcript release factor |
| chr9_+_104063376 | 0.21 |
ENSMUST00000120854.1
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr9_-_44767792 | 0.20 |
ENSMUST00000034607.9
|
Arcn1
|
archain 1 |
| chr11_+_3514861 | 0.20 |
ENSMUST00000094469.4
|
Selm
|
selenoprotein M |
| chr10_+_85102627 | 0.20 |
ENSMUST00000095383.4
|
AI597468
|
expressed sequence AI597468 |
| chr1_+_75479529 | 0.19 |
ENSMUST00000113575.2
ENSMUST00000148980.1 ENSMUST00000050899.6 |
Tmem198
|
transmembrane protein 198 |
| chr7_-_118491912 | 0.18 |
ENSMUST00000178344.1
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
| chr17_-_47833169 | 0.18 |
ENSMUST00000131971.1
ENSMUST00000129360.1 ENSMUST00000113280.1 ENSMUST00000132125.1 |
Mdfi
|
MyoD family inhibitor |
| chr3_+_96697100 | 0.18 |
ENSMUST00000107077.3
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr14_+_26638074 | 0.17 |
ENSMUST00000022429.2
|
Arf4
|
ADP-ribosylation factor 4 |
| chr15_-_76918010 | 0.17 |
ENSMUST00000048854.7
|
Zfp647
|
zinc finger protein 647 |
| chr7_-_122067263 | 0.16 |
ENSMUST00000033159.3
|
Ears2
|
glutamyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr18_+_53176345 | 0.16 |
ENSMUST00000037850.5
|
Snx2
|
sorting nexin 2 |
| chr6_+_70844499 | 0.16 |
ENSMUST00000034093.8
ENSMUST00000162950.1 |
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr7_-_29232478 | 0.15 |
ENSMUST00000085818.4
|
Kcnk6
|
potassium inwardly-rectifying channel, subfamily K, member 6 |
| chr2_-_79428891 | 0.15 |
ENSMUST00000143974.1
|
Cerkl
|
ceramide kinase-like |
| chr4_+_137594140 | 0.15 |
ENSMUST00000105840.1
ENSMUST00000055131.6 ENSMUST00000105839.1 ENSMUST00000105838.1 |
Usp48
|
ubiquitin specific peptidase 48 |
| chr8_+_70302518 | 0.15 |
ENSMUST00000066469.7
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr5_+_113735782 | 0.14 |
ENSMUST00000065698.5
|
Ficd
|
FIC domain containing |
| chr17_-_47833256 | 0.14 |
ENSMUST00000152455.1
ENSMUST00000035375.7 |
Mdfi
|
MyoD family inhibitor |
| chr6_-_54593139 | 0.14 |
ENSMUST00000046520.6
|
Fkbp14
|
FK506 binding protein 14 |
| chr17_-_34031544 | 0.14 |
ENSMUST00000025186.8
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_34031684 | 0.14 |
ENSMUST00000169397.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_34031644 | 0.13 |
ENSMUST00000171872.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_-_32541589 | 0.12 |
ENSMUST00000016231.7
|
Fli1
|
Friend leukemia integration 1 |
| chr7_-_144939823 | 0.12 |
ENSMUST00000093962.4
|
Ccnd1
|
cyclin D1 |
| chr17_-_25785324 | 0.12 |
ENSMUST00000150324.1
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr17_-_75551838 | 0.12 |
ENSMUST00000112507.3
|
Fam98a
|
family with sequence similarity 98, member A |
| chr15_+_23036449 | 0.12 |
ENSMUST00000164787.1
|
Cdh18
|
cadherin 18 |
| chr15_-_48791933 | 0.11 |
ENSMUST00000160658.1
ENSMUST00000100670.3 ENSMUST00000162830.1 |
Csmd3
|
CUB and Sushi multiple domains 3 |
| chr1_+_119526125 | 0.11 |
ENSMUST00000183952.1
|
TMEM185B
|
Transmembrane protein 185B |
| chr4_+_43441939 | 0.11 |
ENSMUST00000060864.6
|
Tesk1
|
testis specific protein kinase 1 |
| chr10_+_61680302 | 0.11 |
ENSMUST00000020285.8
|
Sar1a
|
SAR1 gene homolog A (S. cerevisiae) |
| chr6_-_113740675 | 0.10 |
ENSMUST00000032440.4
|
Sec13
|
SEC13 homolog (S. cerevisiae) |
| chr16_+_52031549 | 0.10 |
ENSMUST00000114471.1
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr16_-_11134601 | 0.10 |
ENSMUST00000118362.1
ENSMUST00000118679.1 |
Txndc11
|
thioredoxin domain containing 11 |
| chr4_+_59003121 | 0.10 |
ENSMUST00000095070.3
ENSMUST00000174664.1 |
Dnajc25
Gm20503
|
DnaJ (Hsp40) homolog, subfamily C, member 25 predicted gene 20503 |
| chr9_+_57560934 | 0.10 |
ENSMUST00000045791.9
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr3_+_58525821 | 0.10 |
ENSMUST00000029387.8
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr10_-_119240006 | 0.10 |
ENSMUST00000020315.6
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr8_+_25720054 | 0.10 |
ENSMUST00000068916.8
ENSMUST00000139836.1 |
Ppapdc1b
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr8_+_70302761 | 0.09 |
ENSMUST00000150968.1
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr10_+_81176631 | 0.09 |
ENSMUST00000047864.9
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr7_+_139212974 | 0.09 |
ENSMUST00000016124.8
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr9_-_89705017 | 0.09 |
ENSMUST00000058488.6
|
Tmed3
|
transmembrane emp24 domain containing 3 |
| chr11_-_59228162 | 0.08 |
ENSMUST00000163300.1
ENSMUST00000061242.7 |
Arf1
|
ADP-ribosylation factor 1 |
| chr14_-_75754475 | 0.08 |
ENSMUST00000049168.7
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr4_-_11386394 | 0.08 |
ENSMUST00000155519.1
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_-_127387113 | 0.08 |
ENSMUST00000164345.1
ENSMUST00000049052.3 |
9130019O22Rik
|
RIKEN cDNA 9130019O22 gene |
| chr17_-_35201996 | 0.08 |
ENSMUST00000167924.1
ENSMUST00000025263.8 |
Tnf
|
tumor necrosis factor |
| chr6_-_88518760 | 0.07 |
ENSMUST00000032168.5
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr5_+_143403819 | 0.07 |
ENSMUST00000110731.2
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr17_+_34031787 | 0.07 |
ENSMUST00000044858.8
|
Rxrb
|
retinoid X receptor beta |
| chr10_-_18743691 | 0.07 |
ENSMUST00000019999.5
|
D10Bwg1379e
|
DNA segment, Chr 10, Brigham & Women's Genetics 1379 expressed |
| chr7_+_139213003 | 0.07 |
ENSMUST00000156768.1
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr4_-_137796350 | 0.07 |
ENSMUST00000030551.4
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr15_+_76701455 | 0.06 |
ENSMUST00000019224.7
|
Mfsd3
|
major facilitator superfamily domain containing 3 |
| chr17_+_34032071 | 0.06 |
ENSMUST00000174299.1
ENSMUST00000173554.1 |
Rxrb
|
retinoid X receptor beta |
| chr1_-_93478785 | 0.06 |
ENSMUST00000170883.1
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr5_+_64970069 | 0.06 |
ENSMUST00000031080.8
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chrX_+_56346390 | 0.06 |
ENSMUST00000101560.3
|
Zfp449
|
zinc finger protein 449 |
| chr1_-_172082757 | 0.06 |
ENSMUST00000003550.4
|
Ncstn
|
nicastrin |
| chr4_-_43562397 | 0.06 |
ENSMUST00000030187.7
|
Tln1
|
talin 1 |
| chr19_-_5295397 | 0.06 |
ENSMUST00000025774.9
|
Sf3b2
|
splicing factor 3b, subunit 2 |
| chr7_+_116504409 | 0.06 |
ENSMUST00000183175.1
|
Nucb2
|
nucleobindin 2 |
| chr2_+_15055274 | 0.05 |
ENSMUST00000069870.3
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr7_+_122067164 | 0.05 |
ENSMUST00000033158.4
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr17_-_51831884 | 0.05 |
ENSMUST00000124222.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr7_-_81992277 | 0.05 |
ENSMUST00000026096.7
|
Bnc1
|
basonuclin 1 |
| chr4_-_47474283 | 0.05 |
ENSMUST00000044148.2
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr9_-_103202113 | 0.05 |
ENSMUST00000035157.8
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr19_-_10101501 | 0.05 |
ENSMUST00000025567.7
|
Fads2
|
fatty acid desaturase 2 |
| chr11_-_68853119 | 0.05 |
ENSMUST00000018880.7
|
Ndel1
|
nuclear distribution gene E-like homolog 1 (A. nidulans) |
| chr14_+_26638237 | 0.04 |
ENSMUST00000112318.3
|
Arf4
|
ADP-ribosylation factor 4 |
| chr16_-_11134624 | 0.04 |
ENSMUST00000038424.7
|
Txndc11
|
thioredoxin domain containing 11 |
| chr11_-_68853019 | 0.04 |
ENSMUST00000108672.1
|
Ndel1
|
nuclear distribution gene E-like homolog 1 (A. nidulans) |
| chr5_+_110176640 | 0.04 |
ENSMUST00000112512.1
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr3_-_84582616 | 0.04 |
ENSMUST00000143514.1
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr2_+_80315461 | 0.04 |
ENSMUST00000028392.7
|
Dnajc10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr7_+_116504363 | 0.04 |
ENSMUST00000032895.8
|
Nucb2
|
nucleobindin 2 |
| chr1_+_172082796 | 0.04 |
ENSMUST00000027833.5
|
Copa
|
coatomer protein complex subunit alpha |
| chr1_+_172082509 | 0.04 |
ENSMUST00000135192.1
|
Copa
|
coatomer protein complex subunit alpha |
| chr6_-_82774448 | 0.04 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr4_+_46489248 | 0.04 |
ENSMUST00000030018.4
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr15_-_57892358 | 0.04 |
ENSMUST00000022993.5
|
Derl1
|
Der1-like domain family, member 1 |
| chr13_+_73937799 | 0.04 |
ENSMUST00000099384.2
|
Brd9
|
bromodomain containing 9 |
| chr12_+_69372112 | 0.03 |
ENSMUST00000050063.7
|
Arf6
|
ADP-ribosylation factor 6 |
| chr2_-_157204483 | 0.03 |
ENSMUST00000029170.7
|
Rbl1
|
retinoblastoma-like 1 (p107) |
| chr17_-_25861456 | 0.03 |
ENSMUST00000079461.8
ENSMUST00000176923.1 |
Wdr90
|
WD repeat domain 90 |
| chr3_-_84582476 | 0.03 |
ENSMUST00000107687.2
ENSMUST00000098990.3 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr19_+_8967031 | 0.03 |
ENSMUST00000052248.7
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr2_-_168742100 | 0.03 |
ENSMUST00000109177.1
|
Atp9a
|
ATPase, class II, type 9A |
| chr8_+_71366848 | 0.03 |
ENSMUST00000110053.2
ENSMUST00000110054.1 ENSMUST00000139541.1 |
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr13_+_55321991 | 0.03 |
ENSMUST00000021942.6
|
Prelid1
|
PRELI domain containing 1 |
| chr10_+_21593151 | 0.03 |
ENSMUST00000057341.4
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr2_-_144011202 | 0.03 |
ENSMUST00000016072.5
ENSMUST00000037875.5 |
Rrbp1
|
ribosome binding protein 1 |
| chr8_+_71367210 | 0.02 |
ENSMUST00000019169.7
|
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr2_-_39065505 | 0.02 |
ENSMUST00000039165.8
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr13_-_76098606 | 0.02 |
ENSMUST00000120573.1
|
Arsk
|
arylsulfatase K |
| chrX_+_73787002 | 0.02 |
ENSMUST00000166518.1
|
Ssr4
|
signal sequence receptor, delta |
| chr2_+_32570858 | 0.02 |
ENSMUST00000140592.1
ENSMUST00000028151.6 |
Dpm2
|
dolichol-phosphate (beta-D) mannosyltransferase 2 |
| chr13_+_77708683 | 0.02 |
ENSMUST00000163257.1
ENSMUST00000091459.4 ENSMUST00000099358.3 |
Fam172a
|
family with sequence similarity 172, member A |
| chr5_+_110176868 | 0.02 |
ENSMUST00000139611.1
ENSMUST00000031477.8 |
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr10_+_19356558 | 0.02 |
ENSMUST00000053225.5
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chrX_-_73786888 | 0.01 |
ENSMUST00000052761.8
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD+), gamma |
| chrX_+_73787062 | 0.01 |
ENSMUST00000002090.2
|
Ssr4
|
signal sequence receptor, delta |
| chr16_-_35490873 | 0.01 |
ENSMUST00000023550.7
|
Pdia5
|
protein disulfide isomerase associated 5 |
| chr2_+_31572775 | 0.01 |
ENSMUST00000137889.1
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr11_-_116274102 | 0.01 |
ENSMUST00000106425.3
|
Srp68
|
signal recognition particle 68 |
| chr7_+_45785331 | 0.01 |
ENSMUST00000120005.1
ENSMUST00000123585.1 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr7_+_24904384 | 0.00 |
ENSMUST00000117419.1
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr14_+_62760496 | 0.00 |
ENSMUST00000181344.1
|
4931440J10Rik
|
RIKEN cDNA 4931440J10 gene |
| chr7_+_100706623 | 0.00 |
ENSMUST00000107042.1
|
Fam168a
|
family with sequence similarity 168, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Xbp1_Creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 3.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 1.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.8 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 1.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 0.6 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.4 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.3 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.2 | GO:2000705 | negative regulation of anion channel activity(GO:0010360) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.7 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 1.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.2 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0070417 | negative regulation of translation in response to stress(GO:0032055) cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 1.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.3 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 3.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 2.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |