Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
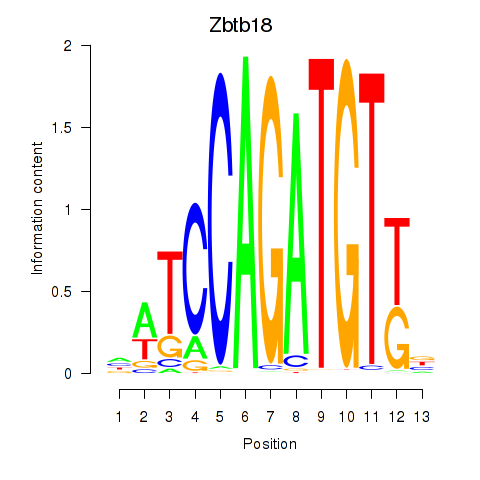
Results for Zbtb18
Z-value: 1.82
Transcription factors associated with Zbtb18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb18
|
ENSMUSG00000063659.6 | zinc finger and BTB domain containing 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb18 | mm10_v2_chr1_+_177445660_177445821 | -0.24 | 1.5e-01 | Click! |
Activity profile of Zbtb18 motif
Sorted Z-values of Zbtb18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_111691002 | 19.74 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr4_-_141825997 | 17.63 |
ENSMUST00000102481.3
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr8_-_105933832 | 13.18 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr17_-_12675833 | 10.32 |
ENSMUST00000024596.8
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr4_-_115496129 | 9.92 |
ENSMUST00000030487.2
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr2_-_28563362 | 9.89 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr1_-_150466165 | 9.82 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr6_-_41035501 | 9.37 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr8_-_24576297 | 8.98 |
ENSMUST00000033953.7
ENSMUST00000121992.1 |
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr14_+_30886521 | 8.96 |
ENSMUST00000168782.1
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr17_-_84682932 | 8.56 |
ENSMUST00000066175.3
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr14_+_30886476 | 8.56 |
ENSMUST00000006703.6
ENSMUST00000078490.5 ENSMUST00000120269.2 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr6_+_37530204 | 7.95 |
ENSMUST00000151256.1
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr6_+_78405148 | 7.78 |
ENSMUST00000023906.2
|
Reg2
|
regenerating islet-derived 2 |
| chr7_-_14446651 | 7.78 |
ENSMUST00000125941.1
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr7_-_14446570 | 7.77 |
ENSMUST00000063509.4
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr6_+_37530173 | 7.45 |
ENSMUST00000040987.7
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr7_-_19699008 | 6.99 |
ENSMUST00000174355.1
ENSMUST00000172983.1 ENSMUST00000174710.1 ENSMUST00000167646.2 ENSMUST00000003066.9 ENSMUST00000174064.1 |
Apoe
|
apolipoprotein E |
| chr1_+_93006328 | 6.92 |
ENSMUST00000059676.4
|
Aqp12
|
aquaporin 12 |
| chr14_+_37054818 | 6.70 |
ENSMUST00000120052.1
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr1_-_139781236 | 6.67 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr7_-_99695628 | 6.58 |
ENSMUST00000145381.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr2_+_43555321 | 6.53 |
ENSMUST00000028223.2
|
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr17_-_28560704 | 6.44 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr7_-_99695572 | 6.33 |
ENSMUST00000137914.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr2_+_43555342 | 6.17 |
ENSMUST00000112826.1
ENSMUST00000050511.6 |
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr7_+_44225430 | 5.90 |
ENSMUST00000075162.3
|
Klk1
|
kallikrein 1 |
| chr12_-_103457195 | 5.47 |
ENSMUST00000044687.6
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr4_-_49506538 | 5.22 |
ENSMUST00000043056.2
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chr17_-_45686120 | 5.09 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr7_-_99695809 | 5.00 |
ENSMUST00000107086.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr12_-_103989950 | 4.95 |
ENSMUST00000120251.2
|
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr17_+_10840361 | 4.88 |
ENSMUST00000097413.3
|
Park2
|
Parkinson disease (autosomal recessive, juvenile) 2, parkin |
| chr9_-_103219823 | 4.87 |
ENSMUST00000168142.1
|
Trf
|
transferrin |
| chr17_-_45685973 | 4.84 |
ENSMUST00000145873.1
|
Tmem63b
|
transmembrane protein 63b |
| chr18_+_45268876 | 4.83 |
ENSMUST00000183850.1
ENSMUST00000066890.7 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr14_-_31640878 | 4.79 |
ENSMUST00000167066.1
ENSMUST00000127204.2 ENSMUST00000022437.8 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr3_-_131303144 | 4.48 |
ENSMUST00000106337.2
|
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr12_-_103989917 | 4.46 |
ENSMUST00000151709.2
ENSMUST00000176246.1 ENSMUST00000074693.7 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr2_+_102706356 | 4.33 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_97417730 | 4.30 |
ENSMUST00000043077.7
|
Thrsp
|
thyroid hormone responsive |
| chr16_+_26581704 | 4.27 |
ENSMUST00000096129.2
ENSMUST00000166294.2 ENSMUST00000174202.1 ENSMUST00000023156.6 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr16_+_91269759 | 4.22 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr4_+_141242850 | 4.17 |
ENSMUST00000138096.1
ENSMUST00000006618.2 ENSMUST00000125392.1 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr17_-_46487641 | 4.16 |
ENSMUST00000047034.8
|
Ttbk1
|
tau tubulin kinase 1 |
| chr1_-_139858684 | 4.14 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr7_+_122671378 | 4.08 |
ENSMUST00000182563.1
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr19_+_34290653 | 4.01 |
ENSMUST00000025691.5
ENSMUST00000112472.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr11_+_99047311 | 3.98 |
ENSMUST00000140772.1
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr19_+_41029275 | 3.92 |
ENSMUST00000051806.4
ENSMUST00000112200.1 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr1_+_160978576 | 3.86 |
ENSMUST00000064725.5
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr1_-_162866502 | 3.84 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr9_+_44066993 | 3.73 |
ENSMUST00000034508.7
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr5_-_92328068 | 3.73 |
ENSMUST00000113093.3
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr3_-_108536466 | 3.71 |
ENSMUST00000048012.6
ENSMUST00000106626.2 ENSMUST00000106625.3 |
5330417C22Rik
|
RIKEN cDNA 5330417C22 gene |
| chr1_+_164796723 | 3.71 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr6_-_35326123 | 3.61 |
ENSMUST00000051176.7
|
Fam180a
|
family with sequence similarity 180, member A |
| chr11_-_115062177 | 3.36 |
ENSMUST00000062787.7
|
Cd300e
|
CD300e antigen |
| chr10_+_63061582 | 3.32 |
ENSMUST00000020266.8
ENSMUST00000178684.1 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr2_-_25500613 | 3.17 |
ENSMUST00000040042.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr9_+_44067072 | 3.14 |
ENSMUST00000177054.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr17_-_32947389 | 3.11 |
ENSMUST00000075253.6
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr7_+_122671401 | 3.11 |
ENSMUST00000182095.1
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr17_-_32947372 | 3.04 |
ENSMUST00000139353.1
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr17_-_7385305 | 3.03 |
ENSMUST00000070059.3
|
Gm9992
|
predicted gene 9992 |
| chr12_-_30373358 | 3.03 |
ENSMUST00000021004.7
|
Sntg2
|
syntrophin, gamma 2 |
| chr4_-_130279205 | 3.02 |
ENSMUST00000120126.2
|
Serinc2
|
serine incorporator 2 |
| chr11_-_68386974 | 3.00 |
ENSMUST00000135141.1
|
Ntn1
|
netrin 1 |
| chr13_+_58806564 | 2.99 |
ENSMUST00000109838.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr8_+_4134733 | 2.99 |
ENSMUST00000130372.1
|
Cd209g
|
CD209g antigen |
| chr4_-_116017854 | 2.97 |
ENSMUST00000049095.5
|
Faah
|
fatty acid amide hydrolase |
| chr7_-_114415128 | 2.91 |
ENSMUST00000163996.1
|
4933406I18Rik
|
RIKEN cDNA 4933406I18 gene |
| chr9_+_119357381 | 2.91 |
ENSMUST00000039610.8
|
Xylb
|
xylulokinase homolog (H. influenzae) |
| chr14_-_63509092 | 2.88 |
ENSMUST00000022522.8
|
Tdh
|
L-threonine dehydrogenase |
| chr4_-_130275542 | 2.86 |
ENSMUST00000154846.1
ENSMUST00000105996.1 |
Serinc2
|
serine incorporator 2 |
| chr1_+_45311538 | 2.83 |
ENSMUST00000087883.6
|
Col3a1
|
collagen, type III, alpha 1 |
| chr8_+_13435459 | 2.82 |
ENSMUST00000167071.1
ENSMUST00000167505.1 |
Tmem255b
|
transmembrane protein 255B |
| chr11_+_118428493 | 2.80 |
ENSMUST00000017590.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr14_-_122913085 | 2.78 |
ENSMUST00000162164.1
ENSMUST00000110679.2 ENSMUST00000038075.5 |
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr4_-_108032069 | 2.76 |
ENSMUST00000106709.2
|
Podn
|
podocan |
| chr4_-_108031938 | 2.72 |
ENSMUST00000106708.1
|
Podn
|
podocan |
| chr17_-_45686214 | 2.70 |
ENSMUST00000113523.2
|
Tmem63b
|
transmembrane protein 63b |
| chr11_+_87582201 | 2.69 |
ENSMUST00000133202.1
|
Sept4
|
septin 4 |
| chr14_+_41105359 | 2.67 |
ENSMUST00000047286.6
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr1_+_88070765 | 2.64 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr5_+_135725713 | 2.62 |
ENSMUST00000127096.1
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr17_-_13131791 | 2.59 |
ENSMUST00000084966.5
|
Unc93a
|
unc-93 homolog A (C. elegans) |
| chr4_-_130275523 | 2.56 |
ENSMUST00000146478.1
|
Serinc2
|
serine incorporator 2 |
| chr19_-_42202150 | 2.51 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr12_-_104013640 | 2.50 |
ENSMUST00000058464.4
|
Serpina9
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chrX_-_8018492 | 2.48 |
ENSMUST00000033503.2
|
Glod5
|
glyoxalase domain containing 5 |
| chr6_-_23132981 | 2.47 |
ENSMUST00000031707.7
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr7_+_43950614 | 2.45 |
ENSMUST00000072204.4
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr5_-_108448882 | 2.36 |
ENSMUST00000031455.3
|
Mfsd7a
|
major facilitator superfamily domain containing 7A |
| chr18_-_32139570 | 2.36 |
ENSMUST00000171765.1
|
Proc
|
protein C |
| chr1_-_132139605 | 2.33 |
ENSMUST00000112362.2
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr1_+_88138364 | 2.31 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr2_-_150904620 | 2.24 |
ENSMUST00000056149.8
|
Abhd12
|
abhydrolase domain containing 12 |
| chr7_-_79848191 | 2.23 |
ENSMUST00000107392.1
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr8_-_4105764 | 2.22 |
ENSMUST00000138439.1
ENSMUST00000145007.1 |
Cd209f
|
CD209f antigen |
| chr5_+_135674572 | 2.21 |
ENSMUST00000153515.1
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr2_-_27247260 | 2.19 |
ENSMUST00000102886.3
ENSMUST00000129975.1 |
Sardh
|
sarcosine dehydrogenase |
| chr12_+_14494561 | 2.19 |
ENSMUST00000052528.3
|
Gm9847
|
predicted pseudogene 9847 |
| chr14_-_68582078 | 2.18 |
ENSMUST00000022641.7
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr3_+_146597077 | 2.14 |
ENSMUST00000029837.7
ENSMUST00000121133.1 |
Uox
|
urate oxidase |
| chr18_-_12862341 | 2.13 |
ENSMUST00000121888.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr4_+_20007938 | 2.12 |
ENSMUST00000125799.1
ENSMUST00000121491.1 |
Ttpa
|
tocopherol (alpha) transfer protein |
| chr5_-_38491948 | 2.11 |
ENSMUST00000129099.1
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr17_-_12868126 | 2.11 |
ENSMUST00000089015.3
|
Mas1
|
MAS1 oncogene |
| chr11_-_74925925 | 2.10 |
ENSMUST00000121738.1
|
Srr
|
serine racemase |
| chr14_+_31641051 | 2.09 |
ENSMUST00000090147.6
|
Btd
|
biotinidase |
| chrX_-_162643575 | 2.06 |
ENSMUST00000101102.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr9_-_36797303 | 2.06 |
ENSMUST00000115086.5
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr7_+_145300806 | 2.04 |
ENSMUST00000033386.5
|
Mrgprf
|
MAS-related GPR, member F |
| chr13_+_30336433 | 2.00 |
ENSMUST00000066412.7
|
Agtr1a
|
angiotensin II receptor, type 1a |
| chr6_+_91156665 | 2.00 |
ENSMUST00000041736.4
|
Hdac11
|
histone deacetylase 11 |
| chr6_-_129237948 | 1.98 |
ENSMUST00000181238.1
ENSMUST00000180379.1 |
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene |
| chr1_-_132139666 | 1.98 |
ENSMUST00000027697.5
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr6_+_91156772 | 1.98 |
ENSMUST00000143621.1
|
Hdac11
|
histone deacetylase 11 |
| chr11_+_118428203 | 1.98 |
ENSMUST00000124861.1
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_+_126950518 | 1.95 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_-_65239092 | 1.90 |
ENSMUST00000156643.1
|
Cobll1
|
Cobl-like 1 |
| chr19_+_43838803 | 1.90 |
ENSMUST00000099413.2
|
Gm10768
|
predicted gene 10768 |
| chr7_+_126950687 | 1.88 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr12_+_85288591 | 1.86 |
ENSMUST00000059341.4
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr7_+_145300889 | 1.86 |
ENSMUST00000117718.1
|
Mrgprf
|
MAS-related GPR, member F |
| chr15_+_31568791 | 1.82 |
ENSMUST00000162532.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr14_+_41151442 | 1.81 |
ENSMUST00000047095.2
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr12_+_29938036 | 1.81 |
ENSMUST00000122328.1
ENSMUST00000118321.1 |
Pxdn
|
peroxidasin homolog (Drosophila) |
| chrX_-_162643629 | 1.81 |
ENSMUST00000112334.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr6_+_70726430 | 1.80 |
ENSMUST00000103410.1
|
Igkc
|
immunoglobulin kappa constant |
| chr4_+_60003438 | 1.80 |
ENSMUST00000107517.1
ENSMUST00000107520.1 |
Mup6
|
major urinary protein 6 |
| chr11_+_101330605 | 1.79 |
ENSMUST00000103105.3
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr11_-_110095974 | 1.77 |
ENSMUST00000100287.2
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr18_+_20665250 | 1.74 |
ENSMUST00000075312.3
|
Ttr
|
transthyretin |
| chr11_-_109722214 | 1.74 |
ENSMUST00000020938.7
|
Fam20a
|
family with sequence similarity 20, member A |
| chr16_+_56204313 | 1.73 |
ENSMUST00000160116.1
ENSMUST00000069936.7 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr12_-_113361232 | 1.70 |
ENSMUST00000103423.1
|
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr6_+_91157373 | 1.69 |
ENSMUST00000155007.1
|
Hdac11
|
histone deacetylase 11 |
| chr19_-_41385070 | 1.69 |
ENSMUST00000059672.7
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr11_+_80154103 | 1.67 |
ENSMUST00000021050.7
|
Adap2
|
ArfGAP with dual PH domains 2 |
| chr9_-_58555129 | 1.67 |
ENSMUST00000165365.1
|
Cd276
|
CD276 antigen |
| chr6_-_50043657 | 1.67 |
ENSMUST00000177892.1
|
Gm21786
|
predicted gene, 21786 |
| chr2_-_168734236 | 1.67 |
ENSMUST00000109175.2
|
Atp9a
|
ATPase, class II, type 9A |
| chr3_+_81932601 | 1.67 |
ENSMUST00000029649.2
|
Ctso
|
cathepsin O |
| chr17_+_79626669 | 1.67 |
ENSMUST00000086570.1
|
4921513D11Rik
|
RIKEN cDNA 4921513D11 gene |
| chr15_+_31568851 | 1.66 |
ENSMUST00000070918.6
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr6_+_72355425 | 1.65 |
ENSMUST00000069695.2
ENSMUST00000132243.1 |
Tmem150a
|
transmembrane protein 150A |
| chr17_+_45686322 | 1.65 |
ENSMUST00000024734.7
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr19_-_5394385 | 1.65 |
ENSMUST00000044527.4
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr5_+_135353295 | 1.64 |
ENSMUST00000111180.2
ENSMUST00000065785.3 |
Trim50
|
tripartite motif-containing 50 |
| chr3_-_88458876 | 1.63 |
ENSMUST00000147200.1
ENSMUST00000169222.1 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr4_-_44066960 | 1.63 |
ENSMUST00000173234.1
ENSMUST00000173274.1 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr17_-_57087729 | 1.62 |
ENSMUST00000071135.5
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr16_-_45693658 | 1.61 |
ENSMUST00000114562.2
ENSMUST00000036617.7 |
Tmprss7
|
transmembrane serine protease 7 |
| chr14_-_20181773 | 1.60 |
ENSMUST00000024011.8
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr17_-_23684019 | 1.60 |
ENSMUST00000085989.5
|
Cldn9
|
claudin 9 |
| chr7_+_100009914 | 1.60 |
ENSMUST00000107084.1
|
Chrdl2
|
chordin-like 2 |
| chr7_-_126898249 | 1.59 |
ENSMUST00000121532.1
ENSMUST00000032926.5 |
Tmem219
|
transmembrane protein 219 |
| chr7_-_98361275 | 1.58 |
ENSMUST00000094161.4
ENSMUST00000164726.1 ENSMUST00000167405.1 |
Tsku
|
tsukushi |
| chr11_+_101331069 | 1.58 |
ENSMUST00000017316.6
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr11_-_68957445 | 1.57 |
ENSMUST00000108671.1
|
Arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr11_-_53773187 | 1.56 |
ENSMUST00000170390.1
|
Gm17334
|
predicted gene, 17334 |
| chr7_+_144175513 | 1.54 |
ENSMUST00000105900.1
|
Shank2
|
SH3/ankyrin domain gene 2 |
| chr4_+_152008803 | 1.53 |
ENSMUST00000097773.3
|
Klhl21
|
kelch-like 21 |
| chr15_-_97020322 | 1.52 |
ENSMUST00000166223.1
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_+_144594054 | 1.52 |
ENSMUST00000136628.1
|
Gm561
|
predicted gene 561 |
| chr3_-_151749877 | 1.52 |
ENSMUST00000029671.7
|
Ifi44
|
interferon-induced protein 44 |
| chr6_+_87428986 | 1.51 |
ENSMUST00000032125.5
|
Bmp10
|
bone morphogenetic protein 10 |
| chr11_-_119086221 | 1.51 |
ENSMUST00000026665.7
|
Cbx4
|
chromobox 4 |
| chr10_+_79970715 | 1.50 |
ENSMUST00000045085.1
|
Grin3b
|
glutamate receptor, ionotropic, NMDA3B |
| chr17_-_10840285 | 1.49 |
ENSMUST00000041463.6
|
Pacrg
|
PARK2 co-regulated |
| chr6_+_72598475 | 1.49 |
ENSMUST00000070597.6
ENSMUST00000176364.1 ENSMUST00000176168.1 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr5_-_24447587 | 1.48 |
ENSMUST00000127194.1
ENSMUST00000115033.1 ENSMUST00000123167.1 ENSMUST00000030799.8 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr11_-_110095937 | 1.48 |
ENSMUST00000106664.3
ENSMUST00000046223.7 ENSMUST00000106662.1 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr7_-_98361310 | 1.47 |
ENSMUST00000165257.1
|
Tsku
|
tsukushi |
| chr2_-_132111440 | 1.47 |
ENSMUST00000128899.1
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr8_-_36732897 | 1.46 |
ENSMUST00000098826.3
|
Dlc1
|
deleted in liver cancer 1 |
| chr15_+_44196135 | 1.46 |
ENSMUST00000038856.6
ENSMUST00000110289.3 |
Trhr
|
thyrotropin releasing hormone receptor |
| chr2_+_69135799 | 1.45 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chr15_+_85017138 | 1.44 |
ENSMUST00000023070.5
|
Upk3a
|
uroplakin 3A |
| chr10_-_18227473 | 1.44 |
ENSMUST00000174592.1
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr7_+_44012672 | 1.43 |
ENSMUST00000048945.4
|
Klk1b26
|
kallikrein 1-related petidase b26 |
| chr5_-_120472763 | 1.43 |
ENSMUST00000052258.7
ENSMUST00000031594.6 |
Sdsl
|
serine dehydratase-like |
| chr11_-_119355484 | 1.43 |
ENSMUST00000100172.2
ENSMUST00000005173.4 |
Sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr11_+_118443471 | 1.43 |
ENSMUST00000133558.1
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr10_-_89621253 | 1.42 |
ENSMUST00000020102.7
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr4_-_44073016 | 1.41 |
ENSMUST00000128439.1
ENSMUST00000140724.2 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr17_+_35439155 | 1.39 |
ENSMUST00000071951.6
ENSMUST00000078205.7 ENSMUST00000116598.3 ENSMUST00000076256.7 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr16_+_23107754 | 1.39 |
ENSMUST00000077605.5
ENSMUST00000115341.3 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr6_+_147531392 | 1.38 |
ENSMUST00000111614.2
|
Ccdc91
|
coiled-coil domain containing 91 |
| chr11_+_49609263 | 1.38 |
ENSMUST00000020617.2
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr9_-_63399216 | 1.37 |
ENSMUST00000168665.1
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr13_+_56438343 | 1.34 |
ENSMUST00000021971.5
|
Slc25a48
|
solute carrier family 25, member 48 |
| chr11_+_44400060 | 1.32 |
ENSMUST00000102796.3
ENSMUST00000170513.1 |
Il12b
|
interleukin 12b |
| chr6_+_17463749 | 1.32 |
ENSMUST00000115443.1
|
Met
|
met proto-oncogene |
| chr7_-_68353182 | 1.30 |
ENSMUST00000123509.1
ENSMUST00000129965.1 |
Gm16158
|
predicted gene 16158 |
| chr11_+_119355551 | 1.28 |
ENSMUST00000050880.7
|
Slc26a11
|
solute carrier family 26, member 11 |
| chr11_-_59182810 | 1.27 |
ENSMUST00000108793.2
|
Gjc2
|
gap junction protein, gamma 2 |
| chr19_+_47865543 | 1.27 |
ENSMUST00000120645.1
|
Gsto2
|
glutathione S-transferase omega 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 4.5 | 17.9 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 4.2 | 12.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 2.9 | 8.6 | GO:1904730 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 2.8 | 8.3 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 2.6 | 10.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 2.3 | 7.0 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 1.2 | 9.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.2 | 9.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.2 | 4.8 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 1.2 | 8.4 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 1.1 | 9.0 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 1.1 | 3.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.0 | 2.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.8 | 2.5 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.8 | 2.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 18.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.8 | 4.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.7 | 2.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.7 | 2.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.7 | 2.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.7 | 4.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.7 | 6.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.7 | 5.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.7 | 4.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.7 | 2.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.6 | 4.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 3.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.6 | 4.8 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.6 | 2.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 1.7 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.6 | 3.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 2.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 1.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.5 | 1.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.5 | 1.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.5 | 1.4 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.5 | 0.9 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 1.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.4 | 5.2 | GO:0019530 | glycine metabolic process(GO:0006544) taurine metabolic process(GO:0019530) |
| 0.4 | 2.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.4 | 5.0 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 2.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.4 | 15.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.4 | 2.7 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.4 | 3.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 3.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 2.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.3 | 2.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 1.0 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.3 | 6.9 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.3 | 1.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.3 | 2.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.3 | 3.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 0.3 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.3 | 4.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 0.9 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.4 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.3 | 1.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.3 | 1.1 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.3 | 1.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 1.1 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.3 | 0.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 2.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.3 | 0.8 | GO:2000848 | positive regulation of corticosteroid hormone secretion(GO:2000848) |
| 0.3 | 1.3 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.3 | 4.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 1.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.5 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 1.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 5.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.2 | 1.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.5 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.2 | 0.8 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 0.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 5.7 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.2 | 0.6 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 1.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 6.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 3.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 0.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.7 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.2 | 1.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 4.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 0.2 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.2 | 3.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 4.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.6 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 20.6 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 1.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 0.5 | GO:0002513 | tolerance induction to self antigen(GO:0002513) endocardial cushion fusion(GO:0003274) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.2 | 0.6 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 2.1 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.1 | 1.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 3.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.7 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 1.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 2.2 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 2.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 1.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 1.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 5.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.5 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 2.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 2.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.8 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.1 | 1.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.8 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.3 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 1.8 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 1.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 3.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 6.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.6 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 3.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 3.9 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.1 | 0.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.6 | GO:0098762 | meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.0 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.0 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.7 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 1.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.3 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 0.9 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.8 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:1903660 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 3.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.9 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.6 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 1.0 | GO:0032008 | activation of MAPKK activity(GO:0000186) positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.2 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.0 | 2.9 | GO:0006304 | DNA modification(GO:0006304) |
| 0.0 | 6.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.7 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 1.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.5 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 1.8 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.6 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 2.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.0 | 1.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.5 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.4 | 7.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.0 | 4.9 | GO:0071797 | LUBAC complex(GO:0071797) Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.7 | 4.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.5 | 4.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 1.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.4 | 4.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 1.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.4 | 1.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.3 | 10.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 1.4 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.3 | 3.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 2.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.3 | 1.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 21.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 0.7 | GO:0016507 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.2 | 1.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 2.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 0.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 1.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 1.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 1.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 8.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 4.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.5 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 7.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.6 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 2.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 91.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 22.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 8.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 5.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 4.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 25.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.8 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 3.4 | 10.3 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 3.3 | 9.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 3.2 | 12.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 3.0 | 9.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 2.3 | 7.0 | GO:0046911 | metal chelating activity(GO:0046911) |
| 1.7 | 9.9 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 1.3 | 4.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 1.2 | 4.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.2 | 4.8 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 1.1 | 3.4 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 1.1 | 4.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.9 | 17.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.7 | 3.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.7 | 2.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.7 | 4.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 17.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 2.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.7 | 3.5 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.7 | 4.9 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.6 | 3.0 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.6 | 8.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 4.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.6 | 2.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.5 | 4.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.5 | 3.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 1.5 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.5 | 2.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 4.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 1.3 | GO:0030613 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.4 | 2.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.4 | 11.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 3.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.3 | 5.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.7 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.3 | 1.0 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.3 | 1.0 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.3 | 1.3 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 8.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.3 | 60.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 5.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 2.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 10.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.2 | 3.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 3.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.8 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.2 | 2.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 4.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 1.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 2.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 5.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 4.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 0.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.7 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 1.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 6.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 11.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 2.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 5.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0043237 | patched binding(GO:0005113) laminin-1 binding(GO:0043237) |
| 0.1 | 2.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 14.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 2.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.5 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0032029 | myosin tail binding(GO:0032029) |
| 0.1 | 1.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 2.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 2.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.1 | GO:0032564 | CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) dATP binding(GO:0032564) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 5.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 1.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.7 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 2.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 17.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 3.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 48.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 3.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 9.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 3.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 7.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 17.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.2 | 21.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.2 | 15.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.9 | 5.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.8 | 10.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.7 | 20.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 6.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 8.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 4.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 3.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 5.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 1.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 7.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 6.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 4.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 3.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 3.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.8 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |