Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
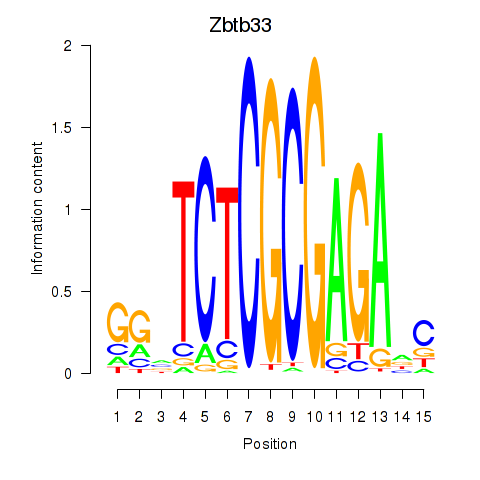
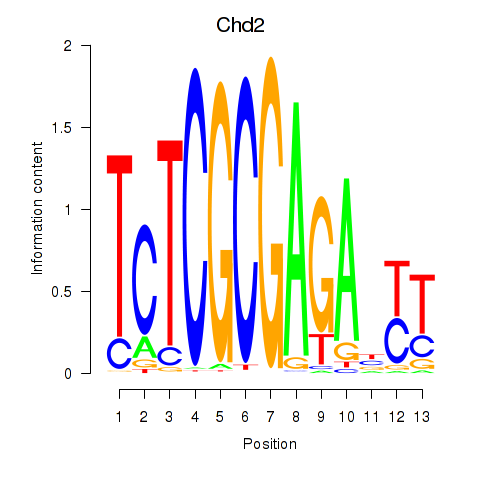
Results for Zbtb33_Chd2
Z-value: 2.68
Transcription factors associated with Zbtb33_Chd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb33
|
ENSMUSG00000048047.3 | zinc finger and BTB domain containing 33 |
|
Chd2
|
ENSMUSG00000078671.4 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb33 | mm10_v2_chrX_+_38189780_38189826 | 0.71 | 1.4e-06 | Click! |
| Chd2 | mm10_v2_chr7_-_73541738_73541758 | 0.56 | 3.5e-04 | Click! |
Activity profile of Zbtb33_Chd2 motif
Sorted Z-values of Zbtb33_Chd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_34833631 | 9.94 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr5_+_45669907 | 7.63 |
ENSMUST00000117396.1
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr3_-_152210017 | 7.60 |
ENSMUST00000029669.3
|
Dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chrX_+_85574018 | 7.23 |
ENSMUST00000048250.3
ENSMUST00000137438.1 ENSMUST00000146063.1 |
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chrX_-_166440671 | 7.19 |
ENSMUST00000049501.8
|
Ofd1
|
oral-facial-digital syndrome 1 gene homolog (human) |
| chr9_+_54863742 | 6.67 |
ENSMUST00000034843.7
|
Ireb2
|
iron responsive element binding protein 2 |
| chr9_+_121719403 | 6.52 |
ENSMUST00000182225.1
|
Nktr
|
natural killer tumor recognition sequence |
| chr3_-_152210032 | 6.50 |
ENSMUST00000144950.1
|
Dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr14_-_105177280 | 6.19 |
ENSMUST00000100327.3
ENSMUST00000022715.7 |
Rbm26
|
RNA binding motif protein 26 |
| chr2_-_156180135 | 5.74 |
ENSMUST00000126992.1
ENSMUST00000146288.1 ENSMUST00000029149.6 ENSMUST00000109587.2 ENSMUST00000109584.1 |
Rbm39
|
RNA binding motif protein 39 |
| chr14_+_32159865 | 5.73 |
ENSMUST00000163336.1
ENSMUST00000169722.1 ENSMUST00000168385.1 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr18_-_34931931 | 5.65 |
ENSMUST00000180351.1
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr11_+_29172890 | 5.62 |
ENSMUST00000102856.2
ENSMUST00000020755.5 |
Smek2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium) |
| chr7_-_131410325 | 5.58 |
ENSMUST00000154602.1
|
Ikzf5
|
IKAROS family zinc finger 5 |
| chr9_-_72491939 | 5.57 |
ENSMUST00000185151.1
ENSMUST00000085358.5 ENSMUST00000184125.1 ENSMUST00000183574.1 ENSMUST00000184831.1 |
Tex9
|
testis expressed gene 9 |
| chr14_-_105177263 | 5.46 |
ENSMUST00000163499.1
|
Rbm26
|
RNA binding motif protein 26 |
| chr17_+_45433823 | 5.38 |
ENSMUST00000181149.1
|
B230354K17Rik
|
RIKEN cDNA B230354K17 gene |
| chr11_-_90638062 | 5.32 |
ENSMUST00000020858.7
ENSMUST00000107875.1 ENSMUST00000107872.1 ENSMUST00000143203.1 |
Stxbp4
|
syntaxin binding protein 4 |
| chr19_+_38931008 | 5.30 |
ENSMUST00000145051.1
|
Hells
|
helicase, lymphoid specific |
| chr16_+_48994185 | 5.29 |
ENSMUST00000117994.1
ENSMUST00000048374.5 |
C330027C09Rik
|
RIKEN cDNA C330027C09 gene |
| chr9_+_121719172 | 5.18 |
ENSMUST00000035112.6
ENSMUST00000182311.1 |
Nktr
|
natural killer tumor recognition sequence |
| chr14_-_78536854 | 5.08 |
ENSMUST00000022593.5
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr7_-_131410495 | 4.97 |
ENSMUST00000121033.1
ENSMUST00000046306.8 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr14_-_78536762 | 4.91 |
ENSMUST00000123853.1
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr6_-_148211996 | 4.71 |
ENSMUST00000126698.1
|
Ergic2
|
ERGIC and golgi 2 |
| chrX_+_23693043 | 4.67 |
ENSMUST00000035766.6
ENSMUST00000101670.2 |
Wdr44
|
WD repeat domain 44 |
| chr3_+_69004969 | 4.67 |
ENSMUST00000136502.1
ENSMUST00000107803.1 |
Smc4
|
structural maintenance of chromosomes 4 |
| chr3_+_69004711 | 4.63 |
ENSMUST00000042901.8
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr1_+_130717320 | 4.35 |
ENSMUST00000049813.4
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae) |
| chr8_-_80739497 | 4.35 |
ENSMUST00000043359.8
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr7_+_133637543 | 4.30 |
ENSMUST00000051169.6
|
2700050L05Rik
|
RIKEN cDNA 2700050L05 gene |
| chr19_+_38930909 | 4.29 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr6_-_148212374 | 4.15 |
ENSMUST00000136008.1
ENSMUST00000032447.4 |
Ergic2
|
ERGIC and golgi 2 |
| chr11_+_98203059 | 4.14 |
ENSMUST00000107539.1
|
Cdk12
|
cyclin-dependent kinase 12 |
| chr11_-_84870812 | 4.09 |
ENSMUST00000168434.1
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chrX_+_13071470 | 3.92 |
ENSMUST00000169594.2
|
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr12_+_116405397 | 3.92 |
ENSMUST00000084828.3
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr2_+_121867083 | 3.89 |
ENSMUST00000089912.5
ENSMUST00000089915.3 |
Casc4
|
cancer susceptibility candidate 4 |
| chr11_-_84870712 | 3.81 |
ENSMUST00000170741.1
ENSMUST00000172405.1 ENSMUST00000100686.3 ENSMUST00000108081.2 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr10_+_11149449 | 3.75 |
ENSMUST00000054814.7
ENSMUST00000159541.1 |
Shprh
|
SNF2 histone linker PHD RING helicase |
| chr11_-_84870646 | 3.72 |
ENSMUST00000018547.2
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr7_+_133637686 | 3.71 |
ENSMUST00000128901.2
|
2700050L05Rik
|
RIKEN cDNA 2700050L05 gene |
| chr7_+_55794146 | 3.70 |
ENSMUST00000032627.3
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr6_+_38551334 | 3.64 |
ENSMUST00000163047.1
ENSMUST00000161538.1 ENSMUST00000057692.4 |
Luc7l2
|
LUC7-like 2 (S. cerevisiae) |
| chr14_-_105176860 | 3.60 |
ENSMUST00000163545.1
|
Rbm26
|
RNA binding motif protein 26 |
| chr3_+_152396664 | 3.59 |
ENSMUST00000089982.4
ENSMUST00000106101.1 |
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr3_-_69004475 | 3.57 |
ENSMUST00000154741.1
ENSMUST00000148031.1 |
Ift80
|
intraflagellar transport 80 |
| chr14_-_57826128 | 3.48 |
ENSMUST00000022536.2
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr11_+_74770908 | 3.48 |
ENSMUST00000128504.1
|
Mettl16
|
methyltransferase like 16 |
| chr13_-_111490028 | 3.46 |
ENSMUST00000091236.4
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chrX_+_151803642 | 3.38 |
ENSMUST00000156616.2
|
Huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr13_+_81783220 | 3.37 |
ENSMUST00000022009.8
|
Cetn3
|
centrin 3 |
| chr5_-_124425572 | 3.36 |
ENSMUST00000168651.1
|
Sbno1
|
sno, strawberry notch homolog 1 (Drosophila) |
| chr1_-_80340311 | 3.34 |
ENSMUST00000164108.1
|
Cul3
|
cullin 3 |
| chr18_+_49832622 | 3.33 |
ENSMUST00000180611.1
|
Dmxl1
|
Dmx-like 1 |
| chr2_+_121866918 | 3.28 |
ENSMUST00000078752.3
ENSMUST00000110586.3 |
Casc4
|
cancer susceptibility candidate 4 |
| chr6_+_38551786 | 3.24 |
ENSMUST00000161227.1
|
Luc7l2
|
LUC7-like 2 (S. cerevisiae) |
| chr3_+_152210458 | 3.23 |
ENSMUST00000166984.1
ENSMUST00000106121.1 |
Fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr1_-_155972887 | 3.17 |
ENSMUST00000138762.1
ENSMUST00000124495.1 |
Cep350
|
centrosomal protein 350 |
| chr9_+_92542223 | 3.15 |
ENSMUST00000070522.7
ENSMUST00000160359.1 |
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr10_+_11149406 | 3.11 |
ENSMUST00000044053.6
|
Shprh
|
SNF2 histone linker PHD RING helicase |
| chr19_+_30030589 | 3.02 |
ENSMUST00000112552.1
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chr11_-_33163072 | 2.92 |
ENSMUST00000093201.6
ENSMUST00000101375.4 ENSMUST00000109354.3 ENSMUST00000075641.3 |
Npm1
|
nucleophosmin 1 |
| chr16_-_48994081 | 2.92 |
ENSMUST00000121869.1
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr1_-_86359455 | 2.87 |
ENSMUST00000027438.6
|
Ncl
|
nucleolin |
| chr2_-_37422869 | 2.85 |
ENSMUST00000112936.1
ENSMUST00000112934.1 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr11_-_77607812 | 2.84 |
ENSMUST00000058496.7
|
Taok1
|
TAO kinase 1 |
| chr16_-_48993931 | 2.83 |
ENSMUST00000114516.1
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr3_-_69004565 | 2.80 |
ENSMUST00000169064.1
|
Ift80
|
intraflagellar transport 80 |
| chr13_-_111490111 | 2.72 |
ENSMUST00000047627.7
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr5_+_123907175 | 2.68 |
ENSMUST00000023869.8
|
Denr
|
density-regulated protein |
| chr9_+_66350465 | 2.68 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr9_+_108517070 | 2.68 |
ENSMUST00000006851.8
ENSMUST00000112155.3 |
Qrich1
|
glutamine-rich 1 |
| chrX_+_13071500 | 2.66 |
ENSMUST00000089302.4
|
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr10_-_88146867 | 2.58 |
ENSMUST00000164121.1
ENSMUST00000164803.1 ENSMUST00000168163.1 ENSMUST00000048518.9 |
Parpbp
|
PARP1 binding protein |
| chr2_+_60209887 | 2.58 |
ENSMUST00000102748.4
ENSMUST00000102747.1 |
March7
|
membrane-associated ring finger (C3HC4) 7 |
| chr3_-_69004503 | 2.58 |
ENSMUST00000107812.1
|
Ift80
|
intraflagellar transport 80 |
| chr2_-_73892588 | 2.55 |
ENSMUST00000154456.1
ENSMUST00000090802.4 ENSMUST00000055833.5 |
Atf2
|
activating transcription factor 2 |
| chr3_+_33800158 | 2.52 |
ENSMUST00000139880.1
ENSMUST00000076916.6 ENSMUST00000142280.1 ENSMUST00000117915.1 ENSMUST00000108210.2 |
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr1_-_128417352 | 2.51 |
ENSMUST00000027602.8
ENSMUST00000064309.7 |
Dars
|
aspartyl-tRNA synthetase |
| chr3_-_86142684 | 2.50 |
ENSMUST00000029722.6
|
Rps3a1
|
ribosomal protein S3A1 |
| chr17_-_42876417 | 2.49 |
ENSMUST00000024709.7
|
Cd2ap
|
CD2-associated protein |
| chr13_+_63815240 | 2.48 |
ENSMUST00000021926.5
ENSMUST00000095724.3 ENSMUST00000143449.1 ENSMUST00000067821.5 |
Ercc6l2
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 like 2 |
| chr1_+_59684949 | 2.48 |
ENSMUST00000027174.3
|
Nop58
|
NOP58 ribonucleoprotein |
| chr1_+_179803376 | 2.47 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chrX_-_37110257 | 2.44 |
ENSMUST00000076265.6
|
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr7_+_90442729 | 2.42 |
ENSMUST00000061767.4
ENSMUST00000107206.1 |
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr15_-_98567630 | 2.41 |
ENSMUST00000012104.6
|
Ccnt1
|
cyclin T1 |
| chr2_-_125123618 | 2.33 |
ENSMUST00000142718.1
ENSMUST00000152367.1 ENSMUST00000067780.3 ENSMUST00000147105.1 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr12_+_111538101 | 2.32 |
ENSMUST00000166123.1
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr2_-_151009364 | 2.32 |
ENSMUST00000109896.1
|
Ninl
|
ninein-like |
| chr2_-_73892619 | 2.30 |
ENSMUST00000112007.1
ENSMUST00000112016.2 |
Atf2
|
activating transcription factor 2 |
| chrX_+_68761875 | 2.26 |
ENSMUST00000114647.1
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr6_+_83326016 | 2.25 |
ENSMUST00000055261.4
|
Mob1a
|
MOB kinase activator 1A |
| chr3_-_122984404 | 2.23 |
ENSMUST00000090379.2
|
Usp53
|
ubiquitin specific peptidase 53 |
| chr1_-_80340480 | 2.21 |
ENSMUST00000163119.1
|
Cul3
|
cullin 3 |
| chr7_+_131410601 | 2.19 |
ENSMUST00000015829.7
ENSMUST00000117518.1 |
Acadsb
|
acyl-Coenzyme A dehydrogenase, short/branched chain |
| chr3_+_88336256 | 2.19 |
ENSMUST00000001451.5
|
Smg5
|
Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr11_+_69070790 | 2.18 |
ENSMUST00000075980.5
ENSMUST00000094081.4 |
Tmem107
|
transmembrane protein 107 |
| chr9_+_64281575 | 2.17 |
ENSMUST00000034964.6
|
Tipin
|
timeless interacting protein |
| chrX_+_166440738 | 2.16 |
ENSMUST00000112192.1
|
Trappc2
|
trafficking protein particle complex 2 |
| chr8_-_31168699 | 2.16 |
ENSMUST00000033983.4
|
Mak16
|
MAK16 homolog (S. cerevisiae) |
| chr6_+_18848571 | 2.15 |
ENSMUST00000056398.8
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chrX_+_42151002 | 2.13 |
ENSMUST00000123245.1
|
Stag2
|
stromal antigen 2 |
| chr4_-_9669068 | 2.11 |
ENSMUST00000078139.6
ENSMUST00000108340.2 ENSMUST00000084915.4 ENSMUST00000108337.1 ENSMUST00000084912.5 ENSMUST00000038564.6 ENSMUST00000146441.1 ENSMUST00000098275.2 |
Asph
|
aspartate-beta-hydroxylase |
| chr4_+_94614483 | 2.10 |
ENSMUST00000030311.4
ENSMUST00000107104.2 |
Ift74
|
intraflagellar transport 74 |
| chr4_+_86575668 | 2.08 |
ENSMUST00000091064.6
|
Rraga
|
Ras-related GTP binding A |
| chr3_-_69044697 | 2.07 |
ENSMUST00000136512.1
ENSMUST00000143454.1 ENSMUST00000107802.1 |
Trim59
|
tripartite motif-containing 59 |
| chr9_-_53667429 | 2.05 |
ENSMUST00000166367.1
ENSMUST00000034529.7 |
Cul5
|
cullin 5 |
| chr1_-_179803625 | 2.04 |
ENSMUST00000027768.7
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr9_-_62811592 | 2.01 |
ENSMUST00000034775.8
|
Fem1b
|
feminization 1 homolog b (C. elegans) |
| chr9_+_14784660 | 2.01 |
ENSMUST00000115632.3
ENSMUST00000147305.1 |
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae) |
| chr7_+_81762947 | 2.01 |
ENSMUST00000133034.1
|
Fam103a1
|
family with sequence similarity 103, member A1 |
| chr2_+_109890846 | 2.00 |
ENSMUST00000028583.7
|
Lin7c
|
lin-7 homolog C (C. elegans) |
| chr7_+_44468051 | 1.99 |
ENSMUST00000118493.1
|
Josd2
|
Josephin domain containing 2 |
| chr5_+_88720855 | 1.98 |
ENSMUST00000113229.1
ENSMUST00000006424.7 |
Mob1b
|
MOB kinase activator 1B |
| chr17_-_45433682 | 1.98 |
ENSMUST00000024727.8
|
Cdc5l
|
cell division cycle 5-like (S. pombe) |
| chr10_+_34282179 | 1.98 |
ENSMUST00000061372.5
|
Tspyl1
|
testis-specific protein, Y-encoded-like 1 |
| chr9_+_25252439 | 1.97 |
ENSMUST00000115272.2
ENSMUST00000165594.2 |
Sept7
|
septin 7 |
| chr11_-_53300373 | 1.91 |
ENSMUST00000020630.7
|
Hspa4
|
heat shock protein 4 |
| chr1_-_160212864 | 1.90 |
ENSMUST00000014370.5
|
Cacybp
|
calcyclin binding protein |
| chr9_+_58582240 | 1.89 |
ENSMUST00000177292.1
ENSMUST00000085651.5 |
Nptn
|
neuroplastin |
| chr7_+_81762925 | 1.88 |
ENSMUST00000042166.4
|
Fam103a1
|
family with sequence similarity 103, member A1 |
| chr4_+_24496434 | 1.86 |
ENSMUST00000108222.2
ENSMUST00000138567.2 ENSMUST00000050446.6 |
Mms22l
|
MMS22-like, DNA repair protein |
| chrX_+_13280970 | 1.85 |
ENSMUST00000000804.6
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked |
| chr6_+_83326071 | 1.84 |
ENSMUST00000038658.8
ENSMUST00000101245.2 |
Mob1a
|
MOB kinase activator 1A |
| chr13_+_58402546 | 1.83 |
ENSMUST00000042450.8
|
Rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr6_+_146577859 | 1.83 |
ENSMUST00000067404.6
ENSMUST00000111663.2 ENSMUST00000058245.4 |
Fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr7_+_44468020 | 1.82 |
ENSMUST00000117324.1
ENSMUST00000120852.1 ENSMUST00000118628.1 |
Josd2
|
Josephin domain containing 2 |
| chr6_-_146577825 | 1.82 |
ENSMUST00000032427.8
|
Asun
|
asunder, spermatogenesis regulator |
| chrX_+_68761890 | 1.82 |
ENSMUST00000071848.6
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr1_-_130715734 | 1.81 |
ENSMUST00000066863.6
ENSMUST00000050406.4 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr2_+_3336159 | 1.79 |
ENSMUST00000115089.1
|
Acbd7
|
acyl-Coenzyme A binding domain containing 7 |
| chr19_-_12796108 | 1.78 |
ENSMUST00000038627.8
|
Zfp91
|
zinc finger protein 91 |
| chr4_+_21727695 | 1.78 |
ENSMUST00000065928.4
|
Ccnc
|
cyclin C |
| chr16_-_17125106 | 1.76 |
ENSMUST00000093336.6
|
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr7_-_113369326 | 1.76 |
ENSMUST00000047091.7
ENSMUST00000119278.1 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chrX_+_68761839 | 1.75 |
ENSMUST00000069731.5
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr5_+_110286306 | 1.73 |
ENSMUST00000007296.5
ENSMUST00000112482.1 |
Pole
|
polymerase (DNA directed), epsilon |
| chr13_-_3611064 | 1.71 |
ENSMUST00000096069.3
|
BC016423
|
cDNA sequence BC016423 |
| chr7_+_44467980 | 1.70 |
ENSMUST00000035844.4
|
Josd2
|
Josephin domain containing 2 |
| chr7_-_34389540 | 1.70 |
ENSMUST00000085585.5
|
Lsm14a
|
LSM14 homolog A (SCD6, S. cerevisiae) |
| chr11_+_116671658 | 1.69 |
ENSMUST00000106378.1
ENSMUST00000144049.1 |
1810032O08Rik
|
RIKEN cDNA 1810032O08 gene |
| chrX_+_37126777 | 1.67 |
ENSMUST00000016553.4
|
Nkap
|
NFKB activating protein |
| chr11_-_107189325 | 1.67 |
ENSMUST00000018577.7
ENSMUST00000106757.1 |
Nol11
|
nucleolar protein 11 |
| chr19_+_30030439 | 1.66 |
ENSMUST00000025739.7
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chrX_-_164076482 | 1.66 |
ENSMUST00000134272.1
|
Siah1b
|
seven in absentia 1B |
| chr3_-_113630068 | 1.65 |
ENSMUST00000092154.3
ENSMUST00000106536.1 ENSMUST00000106535.1 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr14_-_60251473 | 1.60 |
ENSMUST00000041905.6
|
Nupl1
|
nucleoporin like 1 |
| chr3_+_33799791 | 1.59 |
ENSMUST00000099153.3
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr9_+_14784638 | 1.59 |
ENSMUST00000034405.4
|
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae) |
| chr18_+_9958147 | 1.58 |
ENSMUST00000025137.7
|
Thoc1
|
THO complex 1 |
| chr18_+_34331132 | 1.57 |
ENSMUST00000072576.3
ENSMUST00000119329.1 |
Srp19
|
signal recognition particle 19 |
| chr9_-_21963568 | 1.57 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr14_+_99099433 | 1.56 |
ENSMUST00000022650.7
|
Pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr4_+_21727726 | 1.55 |
ENSMUST00000102997.1
ENSMUST00000120679.1 ENSMUST00000108240.2 |
Ccnc
|
cyclin C |
| chr5_+_34660361 | 1.52 |
ENSMUST00000074651.4
ENSMUST00000001112.7 |
Grk4
|
G protein-coupled receptor kinase 4 |
| chr12_-_91384403 | 1.52 |
ENSMUST00000141429.1
|
Cep128
|
centrosomal protein 128 |
| chr18_-_37644185 | 1.52 |
ENSMUST00000066272.4
|
Taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_+_91145103 | 1.50 |
ENSMUST00000094698.1
|
Rbm44
|
RNA binding motif protein 44 |
| chr2_+_69861638 | 1.50 |
ENSMUST00000112260.1
|
Ssb
|
Sjogren syndrome antigen B |
| chr4_+_152039315 | 1.49 |
ENSMUST00000084116.6
ENSMUST00000105663.1 ENSMUST00000103197.3 |
Nol9
|
nucleolar protein 9 |
| chr1_-_24100306 | 1.48 |
ENSMUST00000027337.8
|
Fam135a
|
family with sequence similarity 135, member A |
| chr2_-_151039363 | 1.48 |
ENSMUST00000128627.1
ENSMUST00000066640.4 |
Ninl
Nanp
|
ninein-like N-acetylneuraminic acid phosphatase |
| chr12_+_4082574 | 1.46 |
ENSMUST00000020986.7
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr2_-_34870921 | 1.45 |
ENSMUST00000028225.5
|
Psmd5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr12_+_4082596 | 1.44 |
ENSMUST00000049584.5
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr15_+_6708372 | 1.43 |
ENSMUST00000061656.6
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr1_-_119837613 | 1.41 |
ENSMUST00000064091.5
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chrX_+_109095359 | 1.41 |
ENSMUST00000033598.8
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr9_-_64172879 | 1.38 |
ENSMUST00000176299.1
ENSMUST00000130127.1 ENSMUST00000176794.1 ENSMUST00000177045.1 |
Zwilch
|
zwilch kinetochore protein |
| chr14_-_49066368 | 1.37 |
ENSMUST00000161504.1
|
Exoc5
|
exocyst complex component 5 |
| chr14_+_79451791 | 1.36 |
ENSMUST00000100359.1
|
Zbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr17_+_56040350 | 1.34 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr17_+_33629408 | 1.33 |
ENSMUST00000165504.1
|
Zfp414
|
zinc finger protein 414 |
| chr3_-_144720315 | 1.33 |
ENSMUST00000163279.1
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr16_-_31275277 | 1.33 |
ENSMUST00000060188.7
|
Ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr2_-_119477613 | 1.32 |
ENSMUST00000110808.1
ENSMUST00000049920.7 |
Ino80
|
INO80 homolog (S. cerevisiae) |
| chr4_+_31964081 | 1.32 |
ENSMUST00000037607.4
ENSMUST00000080933.6 ENSMUST00000108183.1 ENSMUST00000108184.2 |
Map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr5_-_34660068 | 1.30 |
ENSMUST00000041364.9
|
Nop14
|
NOP14 nucleolar protein |
| chr8_-_117801903 | 1.29 |
ENSMUST00000034303.1
|
Mphosph6
|
M phase phosphoprotein 6 |
| chr14_-_99099701 | 1.27 |
ENSMUST00000042471.9
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr18_+_24205303 | 1.27 |
ENSMUST00000000430.7
|
Galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chrX_-_164076100 | 1.26 |
ENSMUST00000037928.2
ENSMUST00000071667.2 |
Siah1b
|
seven in absentia 1B |
| chr1_+_105663855 | 1.23 |
ENSMUST00000086721.3
ENSMUST00000039173.6 |
2310035C23Rik
|
RIKEN cDNA 2310035C23 gene |
| chr8_+_46163733 | 1.23 |
ENSMUST00000110376.1
|
4933411K20Rik
|
RIKEN cDNA 4933411K20 gene |
| chr11_+_54522872 | 1.22 |
ENSMUST00000108895.1
ENSMUST00000101206.3 |
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr18_+_35562158 | 1.22 |
ENSMUST00000166793.1
|
Matr3
|
matrin 3 |
| chr16_-_22657182 | 1.22 |
ENSMUST00000023578.7
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr1_-_119837338 | 1.20 |
ENSMUST00000163435.1
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr11_+_54522847 | 1.17 |
ENSMUST00000102743.3
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr9_+_3017408 | 1.17 |
ENSMUST00000099049.3
|
Gm10719
|
predicted gene 10719 |
| chr12_-_3309912 | 1.17 |
ENSMUST00000021001.8
|
Rab10
|
RAB10, member RAS oncogene family |
| chr5_+_4192367 | 1.16 |
ENSMUST00000177258.1
|
Gm9897
|
predicted gene 9897 |
| chr5_+_33658567 | 1.16 |
ENSMUST00000114426.3
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr8_+_84969824 | 1.14 |
ENSMUST00000125893.1
|
Prdx2
|
peroxiredoxin 2 |
| chr5_+_138280516 | 1.14 |
ENSMUST00000048028.8
|
Stag3
|
stromal antigen 3 |
| chr10_+_20312461 | 1.14 |
ENSMUST00000092678.3
ENSMUST00000043881.5 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr5_+_88764983 | 1.13 |
ENSMUST00000031311.9
|
Dck
|
deoxycytidine kinase |
| chr14_+_16365171 | 1.12 |
ENSMUST00000017629.4
|
Top2b
|
topoisomerase (DNA) II beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb33_Chd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 2.3 | 11.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 2.2 | 6.7 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.6 | 6.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.5 | 4.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.4 | 4.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 1.4 | 9.6 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 1.2 | 9.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.1 | 5.6 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 1.0 | 2.9 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.9 | 5.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.9 | 9.7 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.8 | 3.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.8 | 10.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.7 | 3.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.7 | 2.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.7 | 6.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.7 | 2.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.6 | 5.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.6 | 1.7 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.6 | 5.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.6 | 4.5 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.5 | 1.0 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.5 | 2.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.5 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.5 | 1.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.5 | 2.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.5 | 3.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.5 | 4.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.4 | 5.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.4 | 4.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.4 | 3.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.4 | 1.3 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.4 | 11.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 2.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.4 | 2.5 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.4 | 2.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.4 | 2.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.6 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.4 | 4.7 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.4 | 1.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.4 | 3.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 6.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.3 | 1.0 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 | 1.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 1.3 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 | 0.6 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.3 | 0.6 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 1.6 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.3 | 2.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.3 | 2.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 3.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 2.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.3 | 5.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 1.1 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 1.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.3 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.3 | 1.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.3 | 2.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 1.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 2.9 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 2.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 2.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 3.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 1.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 1.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.7 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.5 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 1.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 2.9 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 1.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 1.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.8 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 6.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 1.9 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 3.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.2 | 2.0 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 0.4 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.2 | 0.9 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.2 | 0.7 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 2.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 1.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 2.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 2.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 14.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.6 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 3.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.6 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 6.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.7 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 2.0 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.6 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.4 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 1.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 6.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 4.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.7 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 0.4 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 2.6 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 0.5 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 4.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 2.3 | GO:0072401 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.0 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 3.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 5.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.6 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 2.0 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 1.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 2.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 10.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.9 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 | 0.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.1 | 2.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.6 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 2.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 2.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.5 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 8.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 2.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 11.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 0.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 5.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 | 2.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 1.9 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 1.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.5 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 3.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 11.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 0.3 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) positive regulation of G0 to G1 transition(GO:0070318) positive regulation of hepatocyte proliferation(GO:2000347) |
| 0.1 | 2.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 1.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 3.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.1 | GO:0071492 | response to mycotoxin(GO:0010046) cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.3 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.1 | GO:0043620 | regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 1.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 3.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:2001269 | negative regulation by host of viral transcription(GO:0043922) positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:2000601 | vesicle transport along actin filament(GO:0030050) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.0 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.3 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.7 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.4 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 20.8 | GO:0000796 | condensin complex(GO:0000796) |
| 1.4 | 4.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 1.3 | 3.9 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 1.0 | 13.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.9 | 3.7 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.8 | 5.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.7 | 5.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.7 | 2.0 | GO:0005940 | septin ring(GO:0005940) |
| 0.6 | 1.9 | GO:0031417 | NatC complex(GO:0031417) |
| 0.6 | 1.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.6 | 3.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 2.9 | GO:0001652 | granular component(GO:0001652) |
| 0.5 | 2.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.5 | 2.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 1.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 2.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 1.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 3.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.4 | 2.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 3.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 1.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 2.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 3.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.3 | 8.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 2.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 2.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 4.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 9.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 3.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 2.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 8.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 0.8 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 1.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 0.8 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 0.6 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 2.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 1.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 2.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 5.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.9 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 2.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 3.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 4.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 2.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 2.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 8.4 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 1.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.4 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 5.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 3.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 4.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.6 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 0.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 2.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 6.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 12.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 17.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 2.0 | 9.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.9 | 5.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.6 | 4.7 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 1.3 | 3.9 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.8 | 6.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.8 | 3.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.7 | 2.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.6 | 2.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.5 | 4.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.5 | 3.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 10.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.5 | 1.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.5 | 3.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 4.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 2.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 1.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.4 | 1.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 2.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 14.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 3.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.0 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.3 | 2.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 0.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 2.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 3.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 4.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 11.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 1.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.3 | 1.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 0.8 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.3 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 6.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 8.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 1.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 1.5 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.2 | 0.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 1.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 0.8 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 4.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 3.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 4.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 2.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 1.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 1.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 2.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 2.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 3.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.1 | 1.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.9 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 21.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 3.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.6 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 2.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 8.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 8.8 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.3 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 4.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 4.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 3.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 6.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 15.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.4 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 11.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 2.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 1.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 2.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 11.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 3.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 4.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 1.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 28.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 18.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 2.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 10.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 5.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 10.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 3.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 3.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.3 | 3.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 3.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 6.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 5.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.9 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 9.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 9.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 5.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 5.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 4.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 11.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 3.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 2.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |