Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zbtb4
Z-value: 1.26
Transcription factors associated with Zbtb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb4
|
ENSMUSG00000018750.8 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | mm10_v2_chr11_+_69765899_69765925 | 0.22 | 2.1e-01 | Click! |
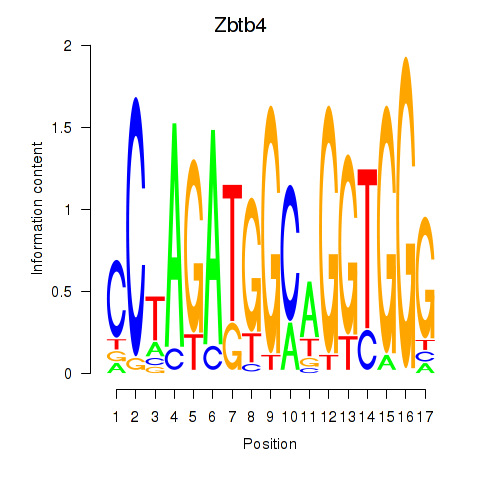
Activity profile of Zbtb4 motif
Sorted Z-values of Zbtb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_90695706 | 11.43 |
ENSMUST00000069960.5
ENSMUST00000117167.1 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr2_-_29869785 | 8.85 |
ENSMUST00000047607.1
|
2600006K01Rik
|
RIKEN cDNA 2600006K01 gene |
| chr7_-_141016892 | 8.59 |
ENSMUST00000081924.3
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr11_-_102107822 | 7.37 |
ENSMUST00000177304.1
ENSMUST00000017455.8 |
Pyy
|
peptide YY |
| chr11_-_3504766 | 7.22 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chrX_-_93832106 | 7.06 |
ENSMUST00000045748.6
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr10_-_62340514 | 6.49 |
ENSMUST00000099691.4
|
Hk1
|
hexokinase 1 |
| chrX_+_73639414 | 6.20 |
ENSMUST00000019701.8
|
Dusp9
|
dual specificity phosphatase 9 |
| chr7_+_142501605 | 5.79 |
ENSMUST00000128294.1
ENSMUST00000146804.1 ENSMUST00000105942.1 ENSMUST00000105947.1 ENSMUST00000105943.1 ENSMUST00000105941.1 ENSMUST00000105946.1 ENSMUST00000105948.1 ENSMUST00000105945.1 ENSMUST00000105944.1 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr12_+_102554966 | 5.74 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr9_-_108190352 | 5.61 |
ENSMUST00000035208.7
|
Bsn
|
bassoon |
| chr2_-_170427828 | 5.41 |
ENSMUST00000013667.2
ENSMUST00000109152.2 ENSMUST00000068137.4 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr8_+_83955507 | 5.34 |
ENSMUST00000005607.8
|
Asf1b
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae) |
| chr2_+_25423234 | 5.28 |
ENSMUST00000134259.1
ENSMUST00000100320.4 |
Fut7
|
fucosyltransferase 7 |
| chr5_-_122050102 | 5.15 |
ENSMUST00000154139.1
|
Cux2
|
cut-like homeobox 2 |
| chr11_-_116076986 | 5.09 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr4_-_156255327 | 4.92 |
ENSMUST00000179919.1
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr6_-_126939524 | 4.88 |
ENSMUST00000144954.1
ENSMUST00000112221.1 ENSMUST00000112220.1 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr2_+_29869484 | 4.34 |
ENSMUST00000047521.6
ENSMUST00000134152.1 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr8_-_105637403 | 4.32 |
ENSMUST00000182046.1
|
Gm5914
|
predicted gene 5914 |
| chr15_+_74563738 | 4.02 |
ENSMUST00000170845.1
|
Bai1
|
brain-specific angiogenesis inhibitor 1 |
| chr16_-_23127702 | 4.02 |
ENSMUST00000115338.1
ENSMUST00000115337.1 ENSMUST00000023598.8 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr15_+_73723131 | 3.81 |
ENSMUST00000165541.1
ENSMUST00000167582.1 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr17_-_25727364 | 3.79 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr7_-_126704179 | 3.72 |
ENSMUST00000106364.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr14_+_31134853 | 3.67 |
ENSMUST00000090212.4
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr17_-_48409729 | 3.67 |
ENSMUST00000160319.1
ENSMUST00000159535.1 ENSMUST00000078800.6 ENSMUST00000046719.7 ENSMUST00000162460.1 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr9_+_59680144 | 3.59 |
ENSMUST00000123914.1
|
Gramd2
|
GRAM domain containing 2 |
| chr8_-_64693027 | 3.55 |
ENSMUST00000048967.7
|
Cpe
|
carboxypeptidase E |
| chr8_+_70501116 | 3.54 |
ENSMUST00000127983.1
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr11_-_116077562 | 3.51 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr5_+_143548700 | 3.47 |
ENSMUST00000169329.1
ENSMUST00000067145.5 ENSMUST00000119488.1 ENSMUST00000118121.1 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr17_+_28801090 | 3.46 |
ENSMUST00000004985.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr10_-_116473875 | 3.34 |
ENSMUST00000068233.4
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr2_-_84822546 | 3.26 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr9_+_75071579 | 3.21 |
ENSMUST00000136731.1
|
Myo5a
|
myosin VA |
| chr17_-_25942821 | 3.19 |
ENSMUST00000148382.1
ENSMUST00000145745.1 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_+_75071386 | 3.15 |
ENSMUST00000155282.2
|
Myo5a
|
myosin VA |
| chr6_+_127887582 | 3.14 |
ENSMUST00000032501.4
|
Tspan11
|
tetraspanin 11 |
| chr6_-_57825055 | 3.12 |
ENSMUST00000127485.1
|
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chrX_-_133898399 | 3.06 |
ENSMUST00000087557.5
|
Tspan6
|
tetraspanin 6 |
| chr7_-_25005895 | 3.05 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr2_+_154200371 | 3.03 |
ENSMUST00000028987.6
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr11_-_116077606 | 3.01 |
ENSMUST00000106450.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr9_+_96196246 | 2.94 |
ENSMUST00000165120.2
ENSMUST00000034982.9 |
Tfdp2
|
transcription factor Dp 2 |
| chr6_-_57825144 | 2.94 |
ENSMUST00000114297.2
|
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr10_+_100015817 | 2.92 |
ENSMUST00000130190.1
ENSMUST00000020129.7 |
Kitl
|
kit ligand |
| chr10_+_128225830 | 2.88 |
ENSMUST00000026455.7
|
Mip
|
major intrinsic protein of eye lens fiber |
| chr7_+_126766397 | 2.84 |
ENSMUST00000032944.7
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr9_+_106203108 | 2.76 |
ENSMUST00000024047.5
|
Twf2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila) |
| chrX_-_133898292 | 2.74 |
ENSMUST00000176718.1
ENSMUST00000176641.1 |
Tspan6
|
tetraspanin 6 |
| chr9_+_108991902 | 2.73 |
ENSMUST00000147989.1
ENSMUST00000051873.8 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr3_-_152340350 | 2.71 |
ENSMUST00000073089.6
ENSMUST00000068243.6 |
Fam73a
|
family with sequence similarity 73, member A |
| chr7_-_126704522 | 2.68 |
ENSMUST00000135087.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chrX_-_111463149 | 2.61 |
ENSMUST00000096348.3
ENSMUST00000113428.2 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr11_-_69895523 | 2.58 |
ENSMUST00000001631.6
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr13_+_35741313 | 2.53 |
ENSMUST00000163595.2
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr11_+_50357759 | 2.45 |
ENSMUST00000052596.2
|
Cby3
|
chibby homolog 3 (Drosophila) |
| chr8_+_39005827 | 2.39 |
ENSMUST00000167992.1
|
Tusc3
|
tumor suppressor candidate 3 |
| chr8_+_39005880 | 2.38 |
ENSMUST00000169034.1
|
Tusc3
|
tumor suppressor candidate 3 |
| chr17_+_6007580 | 2.37 |
ENSMUST00000115784.1
ENSMUST00000115785.1 |
Synj2
|
synaptojanin 2 |
| chr8_+_85299632 | 2.36 |
ENSMUST00000034132.5
ENSMUST00000170141.1 |
Orc6
|
origin recognition complex, subunit 6 |
| chr10_+_45335751 | 2.30 |
ENSMUST00000095715.3
|
Bves
|
blood vessel epicardial substance |
| chr11_-_74590065 | 2.26 |
ENSMUST00000145524.1
ENSMUST00000047488.7 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr9_+_69454066 | 2.23 |
ENSMUST00000134907.1
|
Anxa2
|
annexin A2 |
| chr6_+_112459501 | 2.23 |
ENSMUST00000075477.6
|
Cav3
|
caveolin 3 |
| chr7_+_26061495 | 2.16 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr1_-_134079114 | 2.14 |
ENSMUST00000020692.6
|
Btg2
|
B cell translocation gene 2, anti-proliferative |
| chrX_+_153139941 | 2.13 |
ENSMUST00000039720.4
ENSMUST00000144175.2 |
Rragb
|
Ras-related GTP binding B |
| chr4_-_129440800 | 2.13 |
ENSMUST00000053042.5
ENSMUST00000106046.1 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr8_-_105637350 | 2.12 |
ENSMUST00000182863.1
|
Gm5914
|
predicted gene 5914 |
| chr5_-_117319242 | 2.08 |
ENSMUST00000100834.1
|
Gm10399
|
predicted gene 10399 |
| chr1_+_171388954 | 2.06 |
ENSMUST00000056449.8
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr9_+_119052770 | 1.99 |
ENSMUST00000051386.6
ENSMUST00000074734.6 |
Vill
|
villin-like |
| chr8_-_122678653 | 1.97 |
ENSMUST00000134045.1
|
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr2_+_156840966 | 1.97 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr5_+_110286306 | 1.97 |
ENSMUST00000007296.5
ENSMUST00000112482.1 |
Pole
|
polymerase (DNA directed), epsilon |
| chr16_+_30008657 | 1.96 |
ENSMUST00000181485.1
|
4632428C04Rik
|
RIKEN cDNA 4632428C04 gene |
| chr7_-_24545994 | 1.95 |
ENSMUST00000011776.6
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr1_-_136234113 | 1.94 |
ENSMUST00000120339.1
ENSMUST00000048668.8 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr2_-_73775341 | 1.94 |
ENSMUST00000112024.3
ENSMUST00000166199.1 ENSMUST00000180045.1 |
Chn1
|
chimerin (chimaerin) 1 |
| chr6_+_83237375 | 1.93 |
ENSMUST00000039212.7
ENSMUST00000113899.1 |
Slc4a5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5 |
| chr3_-_57847478 | 1.92 |
ENSMUST00000120289.1
ENSMUST00000066882.8 |
Pfn2
|
profilin 2 |
| chr8_+_106510853 | 1.91 |
ENSMUST00000080797.6
|
Cdh3
|
cadherin 3 |
| chr5_+_149678224 | 1.90 |
ENSMUST00000100404.3
|
B3galtl
|
beta 1,3-galactosyltransferase-like |
| chr9_-_106096776 | 1.89 |
ENSMUST00000121963.1
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr6_+_47244359 | 1.88 |
ENSMUST00000060839.6
|
Cntnap2
|
contactin associated protein-like 2 |
| chr9_-_8042785 | 1.86 |
ENSMUST00000065291.1
|
9230110C19Rik
|
RIKEN cDNA 9230110C19 gene |
| chr3_-_109027600 | 1.85 |
ENSMUST00000171143.1
|
Fam102b
|
family with sequence similarity 102, member B |
| chr19_+_7417586 | 1.83 |
ENSMUST00000159348.1
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr10_-_60219260 | 1.79 |
ENSMUST00000135158.2
|
Chst3
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 3 |
| chr11_-_120990871 | 1.79 |
ENSMUST00000154483.1
|
Csnk1d
|
casein kinase 1, delta |
| chr9_+_106281061 | 1.74 |
ENSMUST00000072206.6
|
Poc1a
|
POC1 centriolar protein homolog A (Chlamydomonas) |
| chr9_+_75071148 | 1.73 |
ENSMUST00000123128.1
|
Myo5a
|
myosin VA |
| chr4_+_130360132 | 1.73 |
ENSMUST00000105994.3
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr11_-_62789445 | 1.72 |
ENSMUST00000054654.6
|
Zfp286
|
zinc finger protein 286 |
| chr16_+_11984581 | 1.70 |
ENSMUST00000170672.2
ENSMUST00000023138.7 |
Shisa9
|
shisa homolog 9 (Xenopus laevis) |
| chr19_+_45047557 | 1.70 |
ENSMUST00000062213.5
ENSMUST00000111954.4 ENSMUST00000084493.6 |
Sfxn3
|
sideroflexin 3 |
| chr4_+_12906838 | 1.68 |
ENSMUST00000143186.1
ENSMUST00000183345.1 |
Triqk
|
triple QxxK/R motif containing |
| chr2_+_32587057 | 1.66 |
ENSMUST00000102818.4
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr11_-_62789402 | 1.62 |
ENSMUST00000108705.1
|
Zfp286
|
zinc finger protein 286 |
| chr6_-_128355826 | 1.62 |
ENSMUST00000001562.6
|
Tulp3
|
tubby-like protein 3 |
| chr4_-_138757578 | 1.59 |
ENSMUST00000030526.6
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr19_-_50678642 | 1.56 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr14_-_34201604 | 1.47 |
ENSMUST00000096019.2
|
Gprin2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr8_+_95081187 | 1.47 |
ENSMUST00000034239.7
|
Katnb1
|
katanin p80 (WD40-containing) subunit B 1 |
| chr18_-_13972617 | 1.46 |
ENSMUST00000025288.7
|
Zfp521
|
zinc finger protein 521 |
| chr8_-_13562397 | 1.46 |
ENSMUST00000151400.2
ENSMUST00000134023.1 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr11_-_69837781 | 1.45 |
ENSMUST00000108634.2
|
Nlgn2
|
neuroligin 2 |
| chr14_+_75016027 | 1.44 |
ENSMUST00000036072.7
|
5031414D18Rik
|
RIKEN cDNA 5031414D18 gene |
| chr4_+_156109971 | 1.43 |
ENSMUST00000072554.6
ENSMUST00000169550.1 ENSMUST00000105576.1 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr2_-_9890026 | 1.42 |
ENSMUST00000130615.1
|
Gata3
|
GATA binding protein 3 |
| chr1_-_171234290 | 1.41 |
ENSMUST00000079957.6
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chrX_+_164373363 | 1.39 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr2_+_30416096 | 1.37 |
ENSMUST00000113601.3
ENSMUST00000113603.3 |
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr2_-_30903255 | 1.36 |
ENSMUST00000102852.3
|
Ptges
|
prostaglandin E synthase |
| chr10_-_81014902 | 1.36 |
ENSMUST00000126317.1
ENSMUST00000092285.3 ENSMUST00000117805.1 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr7_-_35396708 | 1.36 |
ENSMUST00000154597.1
ENSMUST00000032704.5 |
C230052I12Rik
|
RIKEN cDNA C230052I12 gene |
| chr9_-_121495678 | 1.35 |
ENSMUST00000035120.4
|
Cck
|
cholecystokinin |
| chr9_-_37255730 | 1.33 |
ENSMUST00000115068.2
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr18_+_11657349 | 1.32 |
ENSMUST00000047322.6
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr2_+_30416031 | 1.31 |
ENSMUST00000042055.3
|
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr16_+_96235801 | 1.27 |
ENSMUST00000113800.2
|
B3galt5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr2_-_30415509 | 1.27 |
ENSMUST00000134120.1
ENSMUST00000102854.3 |
Crat
|
carnitine acetyltransferase |
| chr8_-_102865853 | 1.23 |
ENSMUST00000076373.6
|
Gm8730
|
predicted pseudogene 8730 |
| chr7_-_125491397 | 1.22 |
ENSMUST00000138616.1
|
Nsmce1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr15_-_85821733 | 1.21 |
ENSMUST00000064370.4
|
Pkdrej
|
polycystic kidney disease (polycystin) and REJ (sperm receptor for egg jelly homolog, sea urchin) |
| chr13_-_55513427 | 1.21 |
ENSMUST00000069929.6
ENSMUST00000069968.6 ENSMUST00000131306.1 ENSMUST00000046246.6 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr12_+_17924294 | 1.17 |
ENSMUST00000169657.1
|
B430203G13Rik
|
RIKEN cDNA B430203G13 gene |
| chr1_+_34678176 | 1.17 |
ENSMUST00000159747.2
|
Arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr4_+_119195353 | 1.17 |
ENSMUST00000106345.2
|
Ccdc23
|
coiled-coil domain containing 23 |
| chr7_+_27553244 | 1.16 |
ENSMUST00000067386.7
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr18_+_42511496 | 1.15 |
ENSMUST00000025375.7
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr3_-_69598822 | 1.14 |
ENSMUST00000061826.1
|
B3galnt1
|
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1 |
| chr1_-_152386589 | 1.10 |
ENSMUST00000162371.1
|
Tsen15
|
tRNA splicing endonuclease 15 homolog (S. cerevisiae) |
| chr19_-_50678485 | 1.09 |
ENSMUST00000111756.3
|
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr2_+_124610573 | 1.08 |
ENSMUST00000103239.3
ENSMUST00000103240.2 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr17_-_29264115 | 1.06 |
ENSMUST00000024802.8
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr7_+_35397046 | 1.05 |
ENSMUST00000079414.5
|
Cep89
|
centrosomal protein 89 |
| chr9_+_37489281 | 1.04 |
ENSMUST00000048604.6
|
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr2_+_130406478 | 1.04 |
ENSMUST00000055421.4
|
Tmem239
|
transmembrane 239 |
| chr2_-_33131645 | 1.03 |
ENSMUST00000133135.1
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr8_-_77724426 | 1.03 |
ENSMUST00000034029.7
|
Ednra
|
endothelin receptor type A |
| chr1_-_36939521 | 1.02 |
ENSMUST00000027290.5
|
Tmem131
|
transmembrane protein 131 |
| chr2_-_30415767 | 0.98 |
ENSMUST00000102855.1
ENSMUST00000028207.6 |
Crat
|
carnitine acetyltransferase |
| chr2_-_84670659 | 0.98 |
ENSMUST00000102646.1
ENSMUST00000102647.3 |
2700094K13Rik
|
RIKEN cDNA 2700094K13 gene |
| chr6_-_49264014 | 0.98 |
ENSMUST00000031841.7
|
Tra2a
|
transformer 2 alpha homolog (Drosophila) |
| chr4_+_6365650 | 0.96 |
ENSMUST00000029912.4
ENSMUST00000103008.5 ENSMUST00000175769.1 ENSMUST00000108374.1 ENSMUST00000140830.1 |
Sdcbp
|
syndecan binding protein |
| chr5_+_117319292 | 0.95 |
ENSMUST00000086464.4
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr13_-_93144557 | 0.95 |
ENSMUST00000062122.3
|
Cmya5
|
cardiomyopathy associated 5 |
| chr1_-_152386675 | 0.94 |
ENSMUST00000015124.8
|
Tsen15
|
tRNA splicing endonuclease 15 homolog (S. cerevisiae) |
| chr6_+_41684414 | 0.92 |
ENSMUST00000031900.5
|
1700034O15Rik
|
RIKEN cDNA 1700034O15 gene |
| chr7_-_142372210 | 0.92 |
ENSMUST00000084412.5
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr11_-_120991039 | 0.92 |
ENSMUST00000070575.7
|
Csnk1d
|
casein kinase 1, delta |
| chr11_-_119111400 | 0.90 |
ENSMUST00000156576.1
|
Gm11754
|
predicted gene 11754 |
| chr3_-_83841767 | 0.88 |
ENSMUST00000029623.9
|
Tlr2
|
toll-like receptor 2 |
| chr11_-_69579320 | 0.87 |
ENSMUST00000048139.5
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chrX_+_41401304 | 0.86 |
ENSMUST00000076349.5
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr14_+_120911177 | 0.85 |
ENSMUST00000032898.7
|
Ipo5
|
importin 5 |
| chr9_-_107635330 | 0.85 |
ENSMUST00000055704.6
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr9_-_21287953 | 0.85 |
ENSMUST00000184326.1
ENSMUST00000038671.3 |
Kri1
|
KRI1 homolog (S. cerevisiae) |
| chr14_+_63436394 | 0.81 |
ENSMUST00000121288.1
|
Fam167a
|
family with sequence similarity 167, member A |
| chr19_+_4081565 | 0.80 |
ENSMUST00000159593.1
|
Cabp2
|
calcium binding protein 2 |
| chr9_-_70657121 | 0.79 |
ENSMUST00000049031.5
|
Fam63b
|
family with sequence similarity 63, member B |
| chr15_+_100761741 | 0.79 |
ENSMUST00000023776.6
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr2_+_124610278 | 0.78 |
ENSMUST00000051419.8
ENSMUST00000077847.5 ENSMUST00000078621.5 ENSMUST00000076335.5 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr14_-_30626196 | 0.77 |
ENSMUST00000112210.3
ENSMUST00000112211.2 ENSMUST00000112208.1 |
Prkcd
|
protein kinase C, delta |
| chr5_+_147957310 | 0.76 |
ENSMUST00000085558.4
ENSMUST00000129092.1 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr7_-_27553138 | 0.76 |
ENSMUST00000127240.1
ENSMUST00000117095.1 ENSMUST00000117611.1 |
Pld3
|
phospholipase D family, member 3 |
| chr2_-_26516620 | 0.76 |
ENSMUST00000132820.1
|
Notch1
|
notch 1 |
| chr11_-_3527916 | 0.75 |
ENSMUST00000020718.4
|
Smtn
|
smoothelin |
| chr6_-_38876163 | 0.71 |
ENSMUST00000161779.1
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr3_+_138143799 | 0.71 |
ENSMUST00000159622.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr14_+_31001383 | 0.71 |
ENSMUST00000168584.1
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr4_+_123105146 | 0.69 |
ENSMUST00000002457.1
|
Bmp8b
|
bone morphogenetic protein 8b |
| chrX_+_20617503 | 0.69 |
ENSMUST00000115375.1
ENSMUST00000115374.1 ENSMUST00000084383.3 |
Rbm10
|
RNA binding motif protein 10 |
| chr15_-_58823530 | 0.68 |
ENSMUST00000072113.5
|
Tmem65
|
transmembrane protein 65 |
| chrX_+_6577259 | 0.68 |
ENSMUST00000089520.2
|
Shroom4
|
shroom family member 4 |
| chr3_+_65528457 | 0.68 |
ENSMUST00000130705.1
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr4_+_129105532 | 0.68 |
ENSMUST00000106064.3
ENSMUST00000030575.8 ENSMUST00000030577.4 |
Tmem54
|
transmembrane protein 54 |
| chr4_-_6454262 | 0.66 |
ENSMUST00000029910.5
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr4_+_125066672 | 0.66 |
ENSMUST00000052183.6
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr7_+_16175085 | 0.64 |
ENSMUST00000176342.1
ENSMUST00000177540.1 |
Meis3
|
Meis homeobox 3 |
| chr9_-_78378725 | 0.61 |
ENSMUST00000034900.7
|
Ooep
|
oocyte expressed protein |
| chr1_-_175692624 | 0.61 |
ENSMUST00000027809.7
|
Opn3
|
opsin 3 |
| chr18_-_77713978 | 0.59 |
ENSMUST00000074653.4
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr4_+_59805829 | 0.57 |
ENSMUST00000030080.6
|
Snx30
|
sorting nexin family member 30 |
| chr11_-_50292302 | 0.57 |
ENSMUST00000059458.4
|
Maml1
|
mastermind like 1 (Drosophila) |
| chr14_+_31001414 | 0.57 |
ENSMUST00000022476.7
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr7_-_142372342 | 0.56 |
ENSMUST00000059223.8
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr18_+_52767994 | 0.56 |
ENSMUST00000025413.7
ENSMUST00000163742.2 ENSMUST00000178011.1 |
Sncaip
|
synuclein, alpha interacting protein (synphilin) |
| chr3_+_152395444 | 0.55 |
ENSMUST00000106103.1
|
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr7_+_16175275 | 0.55 |
ENSMUST00000176506.1
ENSMUST00000002495.11 |
Meis3
|
Meis homeobox 3 |
| chr11_+_75651504 | 0.48 |
ENSMUST00000069057.6
|
Myo1c
|
myosin IC |
| chr9_-_37255403 | 0.48 |
ENSMUST00000161114.1
|
Slc37a2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| chr7_-_132852606 | 0.46 |
ENSMUST00000120425.1
|
Mettl10
|
methyltransferase like 10 |
| chr15_-_101680281 | 0.45 |
ENSMUST00000023786.5
|
Krt6b
|
keratin 6B |
| chr19_-_40612160 | 0.44 |
ENSMUST00000132452.1
ENSMUST00000135795.1 ENSMUST00000025981.8 |
Tctn3
|
tectonic family member 3 |
| chr19_+_41482632 | 0.44 |
ENSMUST00000067795.5
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr13_+_85189433 | 0.43 |
ENSMUST00000165077.1
ENSMUST00000164127.1 ENSMUST00000163600.1 |
Ccnh
|
cyclin H |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 2.0 | 8.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.9 | 5.7 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 1.9 | 5.6 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.8 | 7.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.3 | 11.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.1 | 6.4 | GO:0032796 | uropod organization(GO:0032796) |
| 0.9 | 7.8 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.8 | 6.5 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.7 | 4.3 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.7 | 2.0 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.6 | 5.8 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.6 | 1.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.5 | 8.7 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.5 | 3.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.5 | 1.9 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.5 | 1.4 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.4 | 2.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.4 | 4.4 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.4 | 1.9 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.4 | 2.3 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.4 | 1.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 2.2 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.4 | 2.2 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.4 | 1.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.4 | 3.5 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.3 | 2.8 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.3 | 7.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 3.0 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.3 | 1.3 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.3 | 1.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 1.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 2.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 3.8 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.3 | 3.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 3.0 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.3 | 2.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 6.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 0.9 | GO:0071727 | positive regulation of interleukin-18 production(GO:0032741) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.3 | 1.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 1.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 4.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.3 | 0.8 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 5.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 2.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 2.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 1.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 0.9 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.2 | 3.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 0.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 2.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.2 | 5.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.9 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 1.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 2.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 2.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.0 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 3.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 5.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 2.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.3 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 2.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 3.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.7 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 2.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.1 | 1.4 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.6 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 3.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 2.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.0 | 1.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 3.0 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.7 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.6 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.2 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 1.5 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.9 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0033034 | positive regulation of neutrophil apoptotic process(GO:0033031) positive regulation of myeloid cell apoptotic process(GO:0033034) |
| 0.0 | 0.6 | GO:0035088 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.9 | 5.6 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.1 | 5.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.1 | 11.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 2.9 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.9 | 3.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.9 | 7.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.7 | 2.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.6 | 3.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.6 | 3.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 2.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 2.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 3.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 5.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 4.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 2.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 6.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 1.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 3.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 6.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 1.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 2.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 3.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 6.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 5.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 3.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 8.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 5.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.8 | 7.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.4 | 11.4 | GO:0050786 | Toll-like receptor 4 binding(GO:0035662) RAGE receptor binding(GO:0050786) |
| 1.2 | 3.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 1.0 | 5.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.9 | 5.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.8 | 4.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.7 | 6.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.7 | 7.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.6 | 4.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.6 | 2.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.5 | 2.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 1.8 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.4 | 8.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.4 | 3.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 6.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 4.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 8.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 1.8 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.4 | 1.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 1.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 2.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 6.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 2.7 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 3.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 3.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 3.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) cobalt ion binding(GO:0050897) |
| 0.2 | 2.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 1.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 5.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 2.4 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.2 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.8 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.2 | 3.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 3.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 2.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.9 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 3.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 4.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 3.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.2 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 4.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 6.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 7.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 3.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 5.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 3.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 7.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 6.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 11.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 6.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 2.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 2.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 9.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 5.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 6.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.8 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 9.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 2.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |