Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
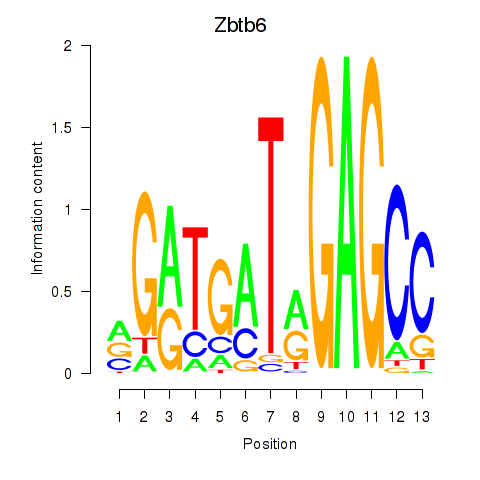
Results for Zbtb6
Z-value: 0.60
Transcription factors associated with Zbtb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb6
|
ENSMUSG00000066798.3 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb6 | mm10_v2_chr2_-_37430835_37430919 | -0.05 | 7.6e-01 | Click! |
Activity profile of Zbtb6 motif
Sorted Z-values of Zbtb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_16863975 | 3.19 |
ENSMUST00000100136.3
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr16_-_16863817 | 2.99 |
ENSMUST00000124890.1
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr8_-_85365341 | 2.93 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_85365317 | 2.93 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr11_-_102897123 | 2.71 |
ENSMUST00000067444.3
|
Gfap
|
glial fibrillary acidic protein |
| chr4_-_87806296 | 2.53 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr11_-_102897146 | 2.48 |
ENSMUST00000077902.4
|
Gfap
|
glial fibrillary acidic protein |
| chr7_-_100855403 | 2.39 |
ENSMUST00000156855.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr17_+_36869567 | 2.31 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr4_-_87806276 | 1.99 |
ENSMUST00000148059.1
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr7_-_141279121 | 1.89 |
ENSMUST00000167790.1
ENSMUST00000046156.6 |
Sct
|
secretin |
| chr11_-_94474088 | 1.76 |
ENSMUST00000107786.1
ENSMUST00000107791.1 ENSMUST00000103166.2 ENSMUST00000107792.1 ENSMUST00000100561.3 ENSMUST00000107793.1 ENSMUST00000107788.1 ENSMUST00000107790.1 ENSMUST00000107789.1 ENSMUST00000107785.1 ENSMUST00000021234.8 |
Cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr6_-_60829826 | 1.66 |
ENSMUST00000163779.1
|
Snca
|
synuclein, alpha |
| chr1_+_134182404 | 1.57 |
ENSMUST00000153856.1
ENSMUST00000082060.3 ENSMUST00000133701.1 ENSMUST00000132873.1 |
Chi3l1
|
chitinase 3-like 1 |
| chr6_+_4755327 | 1.54 |
ENSMUST00000176551.1
|
Peg10
|
paternally expressed 10 |
| chrX_-_53240101 | 1.42 |
ENSMUST00000074861.2
|
Plac1
|
placental specific protein 1 |
| chr1_+_134182150 | 1.34 |
ENSMUST00000156873.1
|
Chi3l1
|
chitinase 3-like 1 |
| chr17_+_25298389 | 1.27 |
ENSMUST00000037453.2
|
Prss34
|
protease, serine, 34 |
| chr11_-_80080928 | 1.26 |
ENSMUST00000103233.3
ENSMUST00000061283.8 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr4_+_107802277 | 1.26 |
ENSMUST00000106733.2
ENSMUST00000030356.3 ENSMUST00000106732.2 ENSMUST00000126573.1 |
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr17_-_45733843 | 1.18 |
ENSMUST00000178179.1
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr3_+_103102604 | 1.13 |
ENSMUST00000173206.1
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr4_-_133887765 | 1.07 |
ENSMUST00000003741.9
ENSMUST00000105894.4 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr4_+_117835387 | 1.04 |
ENSMUST00000169885.1
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_-_76027726 | 1.04 |
ENSMUST00000021207.6
|
Fam101b
|
family with sequence similarity 101, member B |
| chr15_-_101712891 | 1.03 |
ENSMUST00000023709.5
|
Krt5
|
keratin 5 |
| chr4_+_13743424 | 1.03 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_-_98178254 | 1.02 |
ENSMUST00000040971.7
|
Capn5
|
calpain 5 |
| chr6_+_123262107 | 1.02 |
ENSMUST00000032240.2
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr13_+_55369732 | 0.99 |
ENSMUST00000063771.7
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr12_+_111406809 | 0.98 |
ENSMUST00000150384.1
|
A230065H16Rik
|
RIKEN cDNA A230065H16 gene |
| chr6_+_113531675 | 0.95 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr12_-_83951355 | 0.92 |
ENSMUST00000181399.1
|
Gm26571
|
predicted gene, 26571 |
| chr9_-_61976563 | 0.86 |
ENSMUST00000113990.1
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr7_-_38107490 | 0.86 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr11_-_102946688 | 0.85 |
ENSMUST00000057849.5
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr7_-_45136231 | 0.84 |
ENSMUST00000124300.1
ENSMUST00000085377.5 |
Rpl13a
Flt3l
|
ribosomal protein L13A FMS-like tyrosine kinase 3 ligand |
| chr9_-_21760275 | 0.83 |
ENSMUST00000098942.4
|
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chrX_+_50841434 | 0.82 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chr17_+_25366550 | 0.79 |
ENSMUST00000069616.7
|
Tpsb2
|
tryptase beta 2 |
| chr4_-_156200818 | 0.78 |
ENSMUST00000085425.4
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr2_-_126618655 | 0.77 |
ENSMUST00000028838.4
|
Hdc
|
histidine decarboxylase |
| chr9_+_22411515 | 0.77 |
ENSMUST00000058868.7
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr2_-_92371039 | 0.76 |
ENSMUST00000068586.6
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr17_+_28952425 | 0.75 |
ENSMUST00000118762.1
ENSMUST00000057174.8 ENSMUST00000122163.1 ENSMUST00000153831.1 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr2_-_92370999 | 0.75 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr11_-_12026237 | 0.75 |
ENSMUST00000150972.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr5_-_100562836 | 0.74 |
ENSMUST00000097437.2
|
Plac8
|
placenta-specific 8 |
| chr15_+_78913916 | 0.73 |
ENSMUST00000089378.4
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr4_+_127169131 | 0.70 |
ENSMUST00000046659.7
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr12_-_8539545 | 0.69 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr7_+_24547150 | 0.68 |
ENSMUST00000063249.8
|
Xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr8_-_10928449 | 0.66 |
ENSMUST00000040608.3
|
3930402G23Rik
|
RIKEN cDNA 3930402G23 gene |
| chr2_-_92370968 | 0.63 |
ENSMUST00000176774.1
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr6_-_91116785 | 0.62 |
ENSMUST00000113509.1
ENSMUST00000032179.7 |
Nup210
|
nucleoporin 210 |
| chr14_-_54966570 | 0.61 |
ENSMUST00000124930.1
ENSMUST00000134256.1 ENSMUST00000081857.6 ENSMUST00000145322.1 |
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr7_+_28788955 | 0.60 |
ENSMUST00000059857.7
|
Rinl
|
Ras and Rab interactor-like |
| chr17_-_46153517 | 0.60 |
ENSMUST00000171172.1
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr11_+_3488275 | 0.58 |
ENSMUST00000064265.6
|
Pla2g3
|
phospholipase A2, group III |
| chr8_-_105707933 | 0.58 |
ENSMUST00000013299.9
|
Enkd1
|
enkurin domain containing 1 |
| chr3_-_95307132 | 0.57 |
ENSMUST00000015846.2
|
Anxa9
|
annexin A9 |
| chr8_+_75093591 | 0.57 |
ENSMUST00000005548.6
|
Hmox1
|
heme oxygenase (decycling) 1 |
| chr1_+_167001457 | 0.57 |
ENSMUST00000126198.1
|
Fam78b
|
family with sequence similarity 78, member B |
| chr11_-_59035064 | 0.56 |
ENSMUST00000138587.1
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr19_+_33822908 | 0.56 |
ENSMUST00000042061.6
|
Gm5519
|
predicted pseudogene 5519 |
| chr4_-_141053660 | 0.55 |
ENSMUST00000040222.7
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr4_+_124986430 | 0.55 |
ENSMUST00000030687.7
|
Rspo1
|
R-spondin homolog (Xenopus laevis) |
| chr10_-_95673451 | 0.55 |
ENSMUST00000099328.1
|
Anapc15-ps
|
anaphase prompoting complex C subunit 15, pseudogene |
| chr12_-_10900296 | 0.54 |
ENSMUST00000085735.2
|
Pgk1-rs7
|
phosphoglycerate kinase-1, related sequence-7 |
| chr17_+_34850373 | 0.53 |
ENSMUST00000097343.4
ENSMUST00000173357.1 ENSMUST00000173065.1 ENSMUST00000165953.2 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr4_-_141053704 | 0.52 |
ENSMUST00000102491.3
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr2_-_180334665 | 0.51 |
ENSMUST00000015771.2
|
Gata5
|
GATA binding protein 5 |
| chr11_-_71142366 | 0.51 |
ENSMUST00000048514.4
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr17_-_56300286 | 0.50 |
ENSMUST00000097303.1
|
Arrdc5
|
arrestin domain containing 5 |
| chr8_+_83389846 | 0.50 |
ENSMUST00000002259.6
|
Clgn
|
calmegin |
| chr6_-_114042020 | 0.50 |
ENSMUST00000101045.3
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr9_+_108826320 | 0.49 |
ENSMUST00000024238.5
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) |
| chr5_+_47984793 | 0.47 |
ENSMUST00000170109.2
ENSMUST00000174421.1 ENSMUST00000173702.1 ENSMUST00000173107.1 |
Slit2
|
slit homolog 2 (Drosophila) |
| chr8_+_83389878 | 0.47 |
ENSMUST00000109831.2
|
Clgn
|
calmegin |
| chr16_+_18877037 | 0.47 |
ENSMUST00000120532.1
ENSMUST00000004222.7 |
Hira
|
histone cell cycle regulation defective homolog A (S. cerevisiae) |
| chr1_+_64532790 | 0.46 |
ENSMUST00000049932.5
ENSMUST00000087366.4 ENSMUST00000171164.1 |
Creb1
|
cAMP responsive element binding protein 1 |
| chr8_+_85026833 | 0.44 |
ENSMUST00000047281.8
|
2310036O22Rik
|
RIKEN cDNA 2310036O22 gene |
| chr15_+_98108465 | 0.44 |
ENSMUST00000051226.6
|
Pfkm
|
phosphofructokinase, muscle |
| chr3_+_138742195 | 0.44 |
ENSMUST00000029800.2
|
Tspan5
|
tetraspanin 5 |
| chr4_-_133967235 | 0.43 |
ENSMUST00000123234.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr15_+_102990576 | 0.43 |
ENSMUST00000001703.6
|
Hoxc8
|
homeobox C8 |
| chr9_+_66350465 | 0.41 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr17_+_46650328 | 0.40 |
ENSMUST00000043464.7
|
Cul7
|
cullin 7 |
| chr8_-_124021008 | 0.40 |
ENSMUST00000093039.5
|
Taf5l
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr11_-_72361837 | 0.39 |
ENSMUST00000108503.2
|
Tekt1
|
tektin 1 |
| chr11_-_61342821 | 0.38 |
ENSMUST00000134423.1
ENSMUST00000093029.2 |
Slc47a2
|
solute carrier family 47, member 2 |
| chr15_-_79687776 | 0.37 |
ENSMUST00000023061.5
|
Josd1
|
Josephin domain containing 1 |
| chr5_+_47984571 | 0.37 |
ENSMUST00000174313.1
|
Slit2
|
slit homolog 2 (Drosophila) |
| chr9_+_113930934 | 0.37 |
ENSMUST00000084885.5
ENSMUST00000009885.7 |
Ubp1
|
upstream binding protein 1 |
| chr4_-_133967296 | 0.37 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr15_-_95528702 | 0.37 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chr10_-_40142247 | 0.36 |
ENSMUST00000092566.6
|
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr11_-_72411695 | 0.36 |
ENSMUST00000108500.1
ENSMUST00000050226.6 |
Smtnl2
|
smoothelin-like 2 |
| chr17_+_24038144 | 0.36 |
ENSMUST00000059482.5
|
Prss27
|
protease, serine, 27 |
| chr13_+_73317844 | 0.36 |
ENSMUST00000085163.3
|
Gm10263
|
predicted gene 10263 |
| chr7_-_24435491 | 0.34 |
ENSMUST00000080594.6
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr1_+_75549581 | 0.34 |
ENSMUST00000154101.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr3_+_108092789 | 0.34 |
ENSMUST00000058669.8
ENSMUST00000145101.1 |
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr7_-_45136056 | 0.34 |
ENSMUST00000130628.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr7_-_45136102 | 0.34 |
ENSMUST00000125500.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr4_-_141538434 | 0.33 |
ENSMUST00000078886.3
|
Spen
|
SPEN homolog, transcriptional regulator (Drosophila) |
| chr10_+_19712587 | 0.33 |
ENSMUST00000020185.3
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr19_-_31765027 | 0.32 |
ENSMUST00000065067.6
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr15_-_51865448 | 0.31 |
ENSMUST00000022925.8
|
Eif3h
|
eukaryotic translation initiation factor 3, subunit H |
| chr17_-_35115428 | 0.30 |
ENSMUST00000172854.1
ENSMUST00000062657.4 |
Ly6g5b
|
lymphocyte antigen 6 complex, locus G5B |
| chr2_+_139493913 | 0.30 |
ENSMUST00000110083.1
ENSMUST00000047370.2 |
Sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr9_-_45984816 | 0.29 |
ENSMUST00000172450.1
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr6_-_99044414 | 0.29 |
ENSMUST00000177507.1
ENSMUST00000123992.1 |
Foxp1
|
forkhead box P1 |
| chr2_+_31759932 | 0.28 |
ENSMUST00000028190.6
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr19_+_4192129 | 0.28 |
ENSMUST00000046094.4
|
Ppp1ca
|
protein phosphatase 1, catalytic subunit, alpha isoform |
| chr11_-_83429455 | 0.28 |
ENSMUST00000052521.2
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr10_-_75932468 | 0.27 |
ENSMUST00000120281.1
ENSMUST00000000924.6 |
Mmp11
|
matrix metallopeptidase 11 |
| chr4_+_129189760 | 0.26 |
ENSMUST00000106054.2
ENSMUST00000001365.2 |
Yars
|
tyrosyl-tRNA synthetase |
| chr8_+_105605220 | 0.26 |
ENSMUST00000043531.8
|
Fam65a
|
family with sequence similarity 65, member A |
| chr5_-_120588613 | 0.26 |
ENSMUST00000046426.8
|
Tpcn1
|
two pore channel 1 |
| chr7_+_101818306 | 0.26 |
ENSMUST00000008090.9
|
Phox2a
|
paired-like homeobox 2a |
| chr9_-_22135675 | 0.25 |
ENSMUST00000165735.1
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr8_-_116978908 | 0.25 |
ENSMUST00000070577.5
|
1700030J22Rik
|
RIKEN cDNA 1700030J22 gene |
| chr3_+_108093109 | 0.25 |
ENSMUST00000151326.1
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr11_+_67277124 | 0.24 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr14_-_69707546 | 0.23 |
ENSMUST00000118374.1
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr1_-_75264195 | 0.23 |
ENSMUST00000027404.5
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr9_+_107296843 | 0.23 |
ENSMUST00000167072.1
|
Cish
|
cytokine inducible SH2-containing protein |
| chr3_+_96104498 | 0.23 |
ENSMUST00000132980.1
ENSMUST00000138206.1 ENSMUST00000090785.2 ENSMUST00000035519.5 |
Otud7b
|
OTU domain containing 7B |
| chr15_+_39198244 | 0.22 |
ENSMUST00000082054.5
ENSMUST00000042917.9 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_97227364 | 0.22 |
ENSMUST00000046521.7
|
Bcl9
|
B cell CLL/lymphoma 9 |
| chr5_-_107597533 | 0.21 |
ENSMUST00000124140.1
|
Glmn
|
glomulin, FKBP associated protein |
| chr7_+_3289012 | 0.21 |
ENSMUST00000164553.1
|
Myadm
|
myeloid-associated differentiation marker |
| chr5_-_18360384 | 0.21 |
ENSMUST00000074694.5
|
Gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
| chr16_+_58408443 | 0.21 |
ENSMUST00000046663.7
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr11_+_70026815 | 0.20 |
ENSMUST00000135916.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr16_+_38562821 | 0.20 |
ENSMUST00000163948.1
|
Tmem39a
|
transmembrane protein 39a |
| chr4_+_103143052 | 0.20 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr1_-_74588117 | 0.20 |
ENSMUST00000066986.6
|
Zfp142
|
zinc finger protein 142 |
| chr18_-_42262053 | 0.20 |
ENSMUST00000097590.3
|
Lars
|
leucyl-tRNA synthetase |
| chr1_-_74587964 | 0.20 |
ENSMUST00000027315.7
ENSMUST00000113737.1 |
Zfp142
|
zinc finger protein 142 |
| chr4_+_123787857 | 0.20 |
ENSMUST00000053202.5
|
Rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_-_69707493 | 0.20 |
ENSMUST00000121142.1
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr11_-_42182924 | 0.19 |
ENSMUST00000020707.5
ENSMUST00000132971.1 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr11_-_17953861 | 0.18 |
ENSMUST00000076661.6
|
Etaa1
|
Ewing's tumor-associated antigen 1 |
| chr1_-_133131782 | 0.18 |
ENSMUST00000180528.1
|
Gm26616
|
predicted gene, 26616 |
| chr8_-_70439557 | 0.18 |
ENSMUST00000076615.5
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chrX_-_7319186 | 0.18 |
ENSMUST00000115746.1
|
Clcn5
|
chloride channel 5 |
| chr6_-_112489808 | 0.18 |
ENSMUST00000053306.6
|
Oxtr
|
oxytocin receptor |
| chrX_-_7319291 | 0.18 |
ENSMUST00000128319.1
|
Clcn5
|
chloride channel 5 |
| chr17_+_35135695 | 0.17 |
ENSMUST00000174478.1
ENSMUST00000174281.2 ENSMUST00000173550.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr19_-_46573059 | 0.17 |
ENSMUST00000026009.8
|
Arl3
|
ADP-ribosylation factor-like 3 |
| chr2_+_102550012 | 0.17 |
ENSMUST00000028612.7
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr15_-_33405976 | 0.17 |
ENSMUST00000079057.6
|
1700084J12Rik
|
RIKEN cDNA 1700084J12 gene |
| chr4_-_132796361 | 0.17 |
ENSMUST00000045154.5
|
Themis2
|
thymocyte selection associated family member 2 |
| chr9_-_99717259 | 0.16 |
ENSMUST00000112882.2
ENSMUST00000131922.1 |
Cldn18
|
claudin 18 |
| chr16_+_38562806 | 0.16 |
ENSMUST00000171687.1
ENSMUST00000002924.8 |
Tmem39a
|
transmembrane protein 39a |
| chr13_-_40733768 | 0.16 |
ENSMUST00000110193.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr3_-_66981279 | 0.16 |
ENSMUST00000162098.2
|
Shox2
|
short stature homeobox 2 |
| chr11_+_83409655 | 0.15 |
ENSMUST00000175848.1
ENSMUST00000108140.3 |
Rasl10b
|
RAS-like, family 10, member B |
| chr2_-_54085542 | 0.15 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr13_-_98637354 | 0.15 |
ENSMUST00000050389.4
|
Tmem174
|
transmembrane protein 174 |
| chr11_+_3989924 | 0.15 |
ENSMUST00000109981.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr14_-_54913089 | 0.14 |
ENSMUST00000050772.7
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr5_+_73406076 | 0.14 |
ENSMUST00000031040.6
ENSMUST00000065543.6 |
Cwh43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) |
| chr17_-_17624458 | 0.14 |
ENSMUST00000041047.2
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr11_+_114765363 | 0.13 |
ENSMUST00000138804.1
ENSMUST00000084368.5 |
Kif19a
|
kinesin family member 19A |
| chr3_-_92687209 | 0.13 |
ENSMUST00000029524.3
|
Lce1d
|
late cornified envelope 1D |
| chr14_+_54894133 | 0.13 |
ENSMUST00000116476.2
ENSMUST00000022808.7 ENSMUST00000150975.1 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr9_+_108479849 | 0.13 |
ENSMUST00000065014.4
|
Lamb2
|
laminin, beta 2 |
| chr7_-_45136391 | 0.13 |
ENSMUST00000146760.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr10_+_21593151 | 0.13 |
ENSMUST00000057341.4
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr15_+_99126513 | 0.13 |
ENSMUST00000063517.4
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr11_+_74770908 | 0.13 |
ENSMUST00000128504.1
|
Mettl16
|
methyltransferase like 16 |
| chr7_-_84086494 | 0.13 |
ENSMUST00000064174.5
|
9930013L23Rik
|
RIKEN cDNA 9930013L23 gene |
| chr18_-_53744509 | 0.12 |
ENSMUST00000049811.6
|
Cep120
|
centrosomal protein 120 |
| chr1_-_184732444 | 0.12 |
ENSMUST00000174257.1
|
Hlx
|
H2.0-like homeobox |
| chr7_+_127233227 | 0.12 |
ENSMUST00000056232.6
|
Zfp553
|
zinc finger protein 553 |
| chr7_-_21427941 | 0.11 |
ENSMUST00000177741.1
|
Gm6882
|
predicted gene 6882 |
| chr2_-_180104463 | 0.11 |
ENSMUST00000056480.3
|
Hrh3
|
histamine receptor H3 |
| chr14_+_54894573 | 0.11 |
ENSMUST00000141446.1
ENSMUST00000139985.1 ENSMUST00000172557.1 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr1_-_184732616 | 0.11 |
ENSMUST00000048572.6
|
Hlx
|
H2.0-like homeobox |
| chr3_-_65958236 | 0.10 |
ENSMUST00000029416.7
|
Ccnl1
|
cyclin L1 |
| chr7_+_127233044 | 0.10 |
ENSMUST00000106312.3
|
Zfp553
|
zinc finger protein 553 |
| chr10_-_82623190 | 0.10 |
ENSMUST00000183363.1
ENSMUST00000079648.5 ENSMUST00000185168.1 |
1190007I07Rik
|
RIKEN cDNA 1190007I07 gene |
| chr1_+_75546258 | 0.10 |
ENSMUST00000124341.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr8_-_60954726 | 0.10 |
ENSMUST00000110302.1
|
Clcn3
|
chloride channel 3 |
| chr4_-_127061085 | 0.09 |
ENSMUST00000136186.1
ENSMUST00000106099.1 |
Zmym1
|
zinc finger, MYM domain containing 1 |
| chrX_-_162159717 | 0.09 |
ENSMUST00000087085.3
|
Nhs
|
Nance-Horan syndrome (human) |
| chr1_+_134560157 | 0.09 |
ENSMUST00000047714.7
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr8_-_105347885 | 0.09 |
ENSMUST00000014922.4
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_72168142 | 0.09 |
ENSMUST00000013693.6
|
Commd8
|
COMM domain containing 8 |
| chr12_-_3357012 | 0.09 |
ENSMUST00000180719.1
|
Gm26520
|
predicted gene, 26520 |
| chr15_+_58141397 | 0.09 |
ENSMUST00000067563.7
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr5_+_137553517 | 0.08 |
ENSMUST00000136088.1
ENSMUST00000139395.1 ENSMUST00000136565.1 ENSMUST00000149292.1 ENSMUST00000125489.1 |
Actl6b
|
actin-like 6B |
| chr11_+_74770822 | 0.08 |
ENSMUST00000141755.1
ENSMUST00000010698.6 |
Mettl16
|
methyltransferase like 16 |
| chr19_-_23273893 | 0.08 |
ENSMUST00000087556.5
|
Smc5
|
structural maintenance of chromosomes 5 |
| chr14_+_46882854 | 0.07 |
ENSMUST00000022386.8
ENSMUST00000100672.3 |
Samd4
|
sterile alpha motif domain containing 4 |
| chr7_-_45370559 | 0.07 |
ENSMUST00000003971.7
|
Lin7b
|
lin-7 homolog B (C. elegans) |
| chr13_-_21531032 | 0.07 |
ENSMUST00000156674.2
ENSMUST00000110481.2 |
Zkscan8
|
zinc finger with KRAB and SCAN domains 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.9 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.8 | 6.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.6 | 1.9 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of somatostatin secretion(GO:0090274) |
| 0.4 | 1.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.6 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.3 | 4.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 0.8 | GO:0050924 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 | 1.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 1.4 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 0.6 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.2 | 0.7 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.2 | 0.7 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.2 | 0.5 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.2 | 0.8 | GO:0006548 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.1 | 1.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.4 | GO:0021622 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 5.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 2.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.5 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.2 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.6 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.1 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.6 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.6 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.2 | GO:1902606 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 1.3 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0071816 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.4 | 1.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 6.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.9 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 4.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0031464 | Cul2-RING ubiquitin ligase complex(GO:0031462) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 1.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 7.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.7 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 0.4 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 1.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.2 | GO:0030629 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) U6 snRNA 3'-end binding(GO:0030629) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 2.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 4.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:1904315 | GABA-gated chloride ion channel activity(GO:0022851) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 2.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.5 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 1.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |