Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
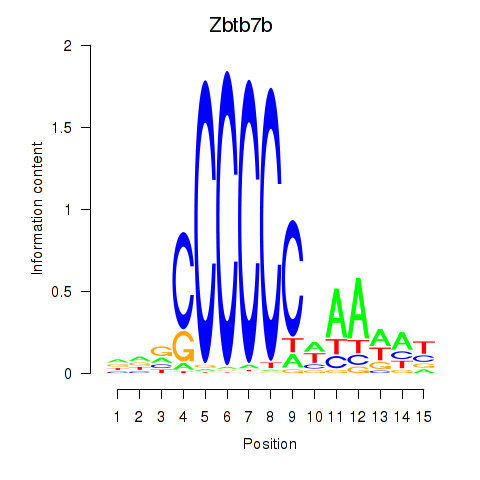
Results for Zbtb7b
Z-value: 0.89
Transcription factors associated with Zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7b
|
ENSMUSG00000028042.9 | zinc finger and BTB domain containing 7B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | mm10_v2_chr3_-_89387132_89387189 | 0.42 | 1.1e-02 | Click! |
Activity profile of Zbtb7b motif
Sorted Z-values of Zbtb7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_84756055 | 5.52 |
ENSMUST00000060397.6
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr6_-_141946960 | 5.25 |
ENSMUST00000042119.5
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr2_+_102658640 | 4.96 |
ENSMUST00000080210.3
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr6_-_141946791 | 4.83 |
ENSMUST00000168119.1
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr7_+_51878967 | 4.52 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr7_+_51879041 | 4.18 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr1_+_167598450 | 3.23 |
ENSMUST00000111386.1
ENSMUST00000111384.1 |
Rxrg
|
retinoid X receptor gamma |
| chr12_-_83597140 | 3.23 |
ENSMUST00000048319.4
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr11_-_113709520 | 3.20 |
ENSMUST00000173655.1
ENSMUST00000100248.4 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chrX_+_36598199 | 3.12 |
ENSMUST00000073339.6
|
Pgrmc1
|
progesterone receptor membrane component 1 |
| chr7_-_141010759 | 3.02 |
ENSMUST00000026565.6
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr3_-_107986360 | 2.85 |
ENSMUST00000066530.6
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chr12_-_84450944 | 2.82 |
ENSMUST00000085192.5
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr1_+_167598384 | 2.77 |
ENSMUST00000015987.3
|
Rxrg
|
retinoid X receptor gamma |
| chr8_+_76902277 | 2.44 |
ENSMUST00000109912.1
ENSMUST00000128862.1 ENSMUST00000109911.1 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr11_-_59449953 | 2.41 |
ENSMUST00000010038.3
ENSMUST00000156146.1 ENSMUST00000132969.1 ENSMUST00000120940.1 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr8_-_117671526 | 2.31 |
ENSMUST00000037955.7
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr11_-_82764303 | 2.23 |
ENSMUST00000021040.3
ENSMUST00000100722.4 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr11_-_59449913 | 2.22 |
ENSMUST00000136436.1
ENSMUST00000150297.1 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr1_+_131970589 | 2.15 |
ENSMUST00000027695.6
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr11_-_69369377 | 2.08 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr9_-_107338305 | 2.08 |
ENSMUST00000118051.1
ENSMUST00000035196.7 |
Hemk1
|
HemK methyltransferase family member 1 |
| chr1_+_78511805 | 1.97 |
ENSMUST00000152111.1
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr13_-_55426769 | 1.97 |
ENSMUST00000170921.1
|
F12
|
coagulation factor XII (Hageman factor) |
| chr1_+_153900572 | 1.93 |
ENSMUST00000139476.1
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr19_+_36554661 | 1.90 |
ENSMUST00000169036.2
ENSMUST00000047247.5 |
Hectd2
|
HECT domain containing 2 |
| chr15_-_50890396 | 1.89 |
ENSMUST00000185183.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr15_-_77533312 | 1.83 |
ENSMUST00000062562.5
|
Apol7c
|
apolipoprotein L 7c |
| chr7_+_18991245 | 1.82 |
ENSMUST00000130268.1
ENSMUST00000059331.8 ENSMUST00000131087.1 |
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr12_-_86892540 | 1.75 |
ENSMUST00000181290.1
|
Gm26698
|
predicted gene, 26698 |
| chr11_+_108920800 | 1.73 |
ENSMUST00000140821.1
|
Axin2
|
axin2 |
| chr7_-_34654342 | 1.73 |
ENSMUST00000108069.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr11_+_108920342 | 1.69 |
ENSMUST00000052915.7
|
Axin2
|
axin2 |
| chr11_-_77519186 | 1.65 |
ENSMUST00000100807.2
|
Gm10392
|
predicted gene 10392 |
| chr15_-_82620907 | 1.62 |
ENSMUST00000109515.1
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr11_-_115187827 | 1.61 |
ENSMUST00000103041.1
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr8_-_41133697 | 1.60 |
ENSMUST00000155055.1
ENSMUST00000059115.6 ENSMUST00000145860.1 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr3_-_89393294 | 1.59 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr11_+_77518566 | 1.59 |
ENSMUST00000147386.1
|
Abhd15
|
abhydrolase domain containing 15 |
| chr15_+_85832301 | 1.58 |
ENSMUST00000146088.1
|
Ttc38
|
tetratricopeptide repeat domain 38 |
| chr11_+_51651179 | 1.58 |
ENSMUST00000170689.1
|
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr11_-_75422586 | 1.57 |
ENSMUST00000138661.1
ENSMUST00000000769.7 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_120617887 | 1.57 |
ENSMUST00000106188.3
ENSMUST00000026129.9 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr7_-_31054815 | 1.57 |
ENSMUST00000071697.4
ENSMUST00000108110.3 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr10_+_34297421 | 1.52 |
ENSMUST00000047935.6
|
Tspyl4
|
TSPY-like 4 |
| chr2_-_160313616 | 1.52 |
ENSMUST00000109475.2
|
Gm826
|
predicted gene 826 |
| chr17_+_36958571 | 1.48 |
ENSMUST00000040177.6
|
Znrd1as
|
Znrd1 antisense |
| chr11_-_75422524 | 1.48 |
ENSMUST00000125982.1
ENSMUST00000137103.1 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr3_-_63964659 | 1.48 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr4_+_43631935 | 1.45 |
ENSMUST00000030191.8
|
Npr2
|
natriuretic peptide receptor 2 |
| chr7_+_28766747 | 1.43 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chr19_+_8839298 | 1.40 |
ENSMUST00000160556.1
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human) |
| chr15_-_76616841 | 1.40 |
ENSMUST00000073428.5
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr17_+_36958623 | 1.38 |
ENSMUST00000173814.1
|
Znrd1as
|
Znrd1 antisense |
| chr19_-_36736653 | 1.34 |
ENSMUST00000087321.2
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C |
| chr3_+_146220955 | 1.33 |
ENSMUST00000039164.2
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr3_+_135825788 | 1.33 |
ENSMUST00000167390.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr17_-_25837082 | 1.32 |
ENSMUST00000183929.1
ENSMUST00000184865.1 ENSMUST00000026831.7 |
Rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila) |
| chr15_-_3303521 | 1.29 |
ENSMUST00000165386.1
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr6_+_88724667 | 1.28 |
ENSMUST00000163271.1
|
Mgll
|
monoglyceride lipase |
| chr15_+_76268076 | 1.27 |
ENSMUST00000074173.3
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr17_+_24850484 | 1.27 |
ENSMUST00000118788.1
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr11_+_70519183 | 1.26 |
ENSMUST00000057685.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr17_+_24850515 | 1.25 |
ENSMUST00000154363.1
ENSMUST00000169200.1 |
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr1_-_171150588 | 1.24 |
ENSMUST00000155798.1
ENSMUST00000081560.4 ENSMUST00000111336.3 |
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chrX_+_142227923 | 1.24 |
ENSMUST00000042329.5
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr1_+_155558112 | 1.22 |
ENSMUST00000080138.6
ENSMUST00000097529.3 ENSMUST00000035560.3 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr8_+_4349588 | 1.19 |
ENSMUST00000110982.1
ENSMUST00000024004.7 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr17_+_33432890 | 1.17 |
ENSMUST00000174088.2
|
Actl9
|
actin-like 9 |
| chr7_-_4522794 | 1.15 |
ENSMUST00000140424.1
|
Tnni3
|
troponin I, cardiac 3 |
| chr17_+_24850742 | 1.12 |
ENSMUST00000149716.2
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr6_-_85902528 | 1.08 |
ENSMUST00000159755.1
|
1700019G17Rik
|
RIKEN cDNA 1700019G17 gene |
| chr7_-_127122226 | 1.07 |
ENSMUST00000032912.5
|
Qprt
|
quinolinate phosphoribosyltransferase |
| chr11_-_62281342 | 1.06 |
ENSMUST00000072916.4
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr17_-_28350747 | 1.06 |
ENSMUST00000080572.7
ENSMUST00000156862.1 |
Tead3
|
TEA domain family member 3 |
| chr17_+_24850654 | 1.05 |
ENSMUST00000130989.1
ENSMUST00000024974.9 |
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr1_-_10009098 | 1.05 |
ENSMUST00000176398.1
ENSMUST00000027049.3 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr4_+_24898080 | 1.03 |
ENSMUST00000029925.3
ENSMUST00000151249.1 |
Ndufaf4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4 |
| chr17_-_28350600 | 1.02 |
ENSMUST00000114799.1
|
Tead3
|
TEA domain family member 3 |
| chr8_-_119635553 | 1.01 |
ENSMUST00000061828.3
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr13_-_48625571 | 1.00 |
ENSMUST00000035824.9
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr6_-_83572429 | 1.00 |
ENSMUST00000068054.7
|
Stambp
|
STAM binding protein |
| chr17_-_32189457 | 0.98 |
ENSMUST00000087721.3
ENSMUST00000162117.1 |
Ephx3
|
epoxide hydrolase 3 |
| chr11_-_42182163 | 0.96 |
ENSMUST00000153147.1
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr6_+_122707489 | 0.96 |
ENSMUST00000112581.1
ENSMUST00000112580.1 ENSMUST00000012540.4 |
Nanog
|
Nanog homeobox |
| chr9_-_22052021 | 0.95 |
ENSMUST00000003501.7
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) |
| chr15_-_76232045 | 0.95 |
ENSMUST00000167754.1
|
Plec
|
plectin |
| chr9_+_68653761 | 0.94 |
ENSMUST00000034766.7
|
Rora
|
RAR-related orphan receptor alpha |
| chr7_-_101837776 | 0.93 |
ENSMUST00000165052.1
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr2_-_173276526 | 0.93 |
ENSMUST00000036248.6
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr8_+_83972951 | 0.91 |
ENSMUST00000005606.6
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr4_+_138395198 | 0.91 |
ENSMUST00000062902.6
|
AB041806
|
hypothetical protein, MNCb-2457 |
| chr11_+_108921648 | 0.90 |
ENSMUST00000144511.1
|
Axin2
|
axin2 |
| chr14_+_11553523 | 0.89 |
ENSMUST00000022264.6
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr4_-_130279205 | 0.88 |
ENSMUST00000120126.2
|
Serinc2
|
serine incorporator 2 |
| chr5_-_146963742 | 0.88 |
ENSMUST00000125217.1
ENSMUST00000110564.1 ENSMUST00000066675.3 ENSMUST00000016654.2 ENSMUST00000110566.1 ENSMUST00000140526.1 |
Mtif3
|
mitochondrial translational initiation factor 3 |
| chr19_-_6992478 | 0.87 |
ENSMUST00000025915.5
|
Dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr11_-_83302586 | 0.87 |
ENSMUST00000176374.1
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr16_+_23429133 | 0.86 |
ENSMUST00000038730.6
|
Rtp1
|
receptor transporter protein 1 |
| chr14_-_16249154 | 0.84 |
ENSMUST00000148121.1
ENSMUST00000112624.1 |
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr11_+_88068242 | 0.82 |
ENSMUST00000018521.4
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chr8_+_35587780 | 0.82 |
ENSMUST00000037666.5
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr16_+_78301458 | 0.80 |
ENSMUST00000023572.7
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr8_+_13037802 | 0.78 |
ENSMUST00000152034.1
ENSMUST00000128418.1 |
F10
|
coagulation factor X |
| chr3_+_52268337 | 0.78 |
ENSMUST00000053764.5
|
Foxo1
|
forkhead box O1 |
| chr19_-_6909599 | 0.77 |
ENSMUST00000173091.1
|
Prdx5
|
peroxiredoxin 5 |
| chr6_-_134792596 | 0.76 |
ENSMUST00000100857.3
|
Dusp16
|
dual specificity phosphatase 16 |
| chr15_-_10558644 | 0.75 |
ENSMUST00000166039.1
ENSMUST00000022857.7 |
Ttc23l
|
tetratricopeptide repeat domain 23-like |
| chr17_+_6978860 | 0.74 |
ENSMUST00000089119.5
ENSMUST00000179728.1 |
Rnaset2b
|
ribonuclease T2B |
| chr15_-_76243401 | 0.73 |
ENSMUST00000165738.1
ENSMUST00000075689.6 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr11_+_70525361 | 0.70 |
ENSMUST00000018430.6
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr7_-_28766469 | 0.69 |
ENSMUST00000085851.5
ENSMUST00000032815.4 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr5_+_102768771 | 0.68 |
ENSMUST00000112852.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_-_44135816 | 0.68 |
ENSMUST00000026218.5
|
Cwf19l1
|
CWF19-like 1, cell cycle control (S. pombe) |
| chr7_-_4971168 | 0.67 |
ENSMUST00000133272.1
|
Gm1078
|
predicted gene 1078 |
| chr7_+_140847801 | 0.66 |
ENSMUST00000026555.5
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr5_+_36464998 | 0.66 |
ENSMUST00000031099.3
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr7_-_30598863 | 0.65 |
ENSMUST00000108150.1
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr8_-_23257009 | 0.65 |
ENSMUST00000121783.1
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr11_+_88718442 | 0.64 |
ENSMUST00000138007.1
|
C030037D09Rik
|
RIKEN cDNA C030037D09 gene |
| chr6_+_108213086 | 0.64 |
ENSMUST00000032192.6
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr8_-_23257043 | 0.64 |
ENSMUST00000051094.6
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr6_-_134887783 | 0.64 |
ENSMUST00000066107.6
|
Gpr19
|
G protein-coupled receptor 19 |
| chr6_+_41538218 | 0.63 |
ENSMUST00000103291.1
|
Trbc1
|
T cell receptor beta, constant region 1 |
| chr19_-_6910088 | 0.63 |
ENSMUST00000025904.5
|
Prdx5
|
peroxiredoxin 5 |
| chr2_-_151668533 | 0.62 |
ENSMUST00000180195.1
ENSMUST00000096439.3 |
Rad21l
|
RAD21-like (S. pombe) |
| chr9_-_110117303 | 0.60 |
ENSMUST00000136969.1
|
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr3_+_135825648 | 0.59 |
ENSMUST00000180196.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr11_+_82388900 | 0.58 |
ENSMUST00000054245.4
ENSMUST00000092852.2 |
Tmem132e
|
transmembrane protein 132E |
| chr17_+_34325658 | 0.58 |
ENSMUST00000050325.8
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr17_+_85620816 | 0.57 |
ENSMUST00000175898.2
|
Six3
|
sine oculis-related homeobox 3 |
| chrX_+_7762652 | 0.57 |
ENSMUST00000077680.3
ENSMUST00000079542.6 ENSMUST00000115679.1 ENSMUST00000137467.1 |
Tfe3
|
transcription factor E3 |
| chr9_-_121839460 | 0.56 |
ENSMUST00000135986.2
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr13_-_67755192 | 0.56 |
ENSMUST00000144183.1
|
Zfp85-rs1
|
zinc finger protein 85, related sequence 1 |
| chr11_-_115276973 | 0.54 |
ENSMUST00000021078.2
|
Fdxr
|
ferredoxin reductase |
| chr5_+_139543889 | 0.54 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr2_+_163506808 | 0.54 |
ENSMUST00000143911.1
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr5_-_72974702 | 0.53 |
ENSMUST00000043711.8
|
Gm10135
|
predicted gene 10135 |
| chr11_+_75513540 | 0.53 |
ENSMUST00000042808.6
ENSMUST00000118243.1 |
Scarf1
|
scavenger receptor class F, member 1 |
| chr17_+_35236556 | 0.53 |
ENSMUST00000068261.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr9_-_91365778 | 0.53 |
ENSMUST00000065360.3
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr3_+_34649987 | 0.53 |
ENSMUST00000099151.2
|
Sox2
|
SRY-box containing gene 2 |
| chr11_+_59450040 | 0.52 |
ENSMUST00000045279.6
ENSMUST00000108777.3 |
Jmjd4
|
jumonji domain containing 4 |
| chrX_-_101419788 | 0.52 |
ENSMUST00000117901.1
ENSMUST00000120201.1 ENSMUST00000117637.1 ENSMUST00000134005.1 ENSMUST00000121520.1 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr7_+_140847816 | 0.51 |
ENSMUST00000106049.1
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr7_-_141214080 | 0.51 |
ENSMUST00000026573.5
ENSMUST00000170841.1 |
1600016N20Rik
|
RIKEN cDNA 1600016N20 gene |
| chr2_-_103797617 | 0.50 |
ENSMUST00000028607.6
|
Caprin1
|
cell cycle associated protein 1 |
| chr4_+_152338887 | 0.50 |
ENSMUST00000005175.4
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr14_+_16249259 | 0.50 |
ENSMUST00000022310.6
|
Ngly1
|
N-glycanase 1 |
| chr7_-_78783026 | 0.50 |
ENSMUST00000032841.5
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr9_-_44417983 | 0.49 |
ENSMUST00000053286.7
|
Ccdc84
|
coiled-coil domain containing 84 |
| chr17_-_36958437 | 0.47 |
ENSMUST00000113669.2
|
Znrd1
|
zinc ribbon domain containing, 1 |
| chr6_-_52191695 | 0.46 |
ENSMUST00000101395.2
|
Hoxa4
|
homeobox A4 |
| chr14_-_50897456 | 0.46 |
ENSMUST00000170855.1
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr18_+_48045329 | 0.45 |
ENSMUST00000076155.4
|
Gm5506
|
predicted gene 5506 |
| chr11_+_101009452 | 0.45 |
ENSMUST00000044721.6
ENSMUST00000103110.3 ENSMUST00000168757.2 |
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr8_-_119635346 | 0.44 |
ENSMUST00000164382.1
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr1_-_74935549 | 0.44 |
ENSMUST00000094844.3
|
Ccdc108
|
coiled-coil domain containing 108 |
| chr16_-_44333135 | 0.44 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_-_43771009 | 0.44 |
ENSMUST00000053931.1
|
Olfr159
|
olfactory receptor 159 |
| chr16_+_20673264 | 0.43 |
ENSMUST00000154950.1
ENSMUST00000115461.1 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr7_-_4970961 | 0.43 |
ENSMUST00000144863.1
|
Gm1078
|
predicted gene 1078 |
| chr4_-_130308674 | 0.43 |
ENSMUST00000097865.1
|
Gm10570
|
predicted gene 10570 |
| chr11_+_83302641 | 0.43 |
ENSMUST00000176430.1
ENSMUST00000065692.7 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr6_+_14901344 | 0.42 |
ENSMUST00000115477.1
|
Foxp2
|
forkhead box P2 |
| chr2_-_109278274 | 0.42 |
ENSMUST00000081631.3
|
Mettl15
|
methyltransferase like 15 |
| chr11_-_75454656 | 0.42 |
ENSMUST00000173320.1
|
Wdr81
|
WD repeat domain 81 |
| chr11_-_17211504 | 0.41 |
ENSMUST00000020317.7
|
Pno1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr7_-_127725616 | 0.41 |
ENSMUST00000076091.2
|
Ctf2
|
cardiotrophin 2 |
| chr6_+_67115671 | 0.41 |
ENSMUST00000181304.1
|
A430010J10Rik
|
RIKEN cDNA A430010J10 gene |
| chr2_-_155945282 | 0.40 |
ENSMUST00000040162.2
|
Gdf5
|
growth differentiation factor 5 |
| chr7_+_78783119 | 0.40 |
ENSMUST00000032840.4
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr11_+_82911253 | 0.40 |
ENSMUST00000164945.1
ENSMUST00000018989.7 |
Unc45b
|
unc-45 homolog B (C. elegans) |
| chr11_-_22982090 | 0.39 |
ENSMUST00000160826.1
ENSMUST00000093270.5 ENSMUST00000071068.8 ENSMUST00000159081.1 |
RP23-242C19.7
Commd1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (B3gnt2), transcript variant 2, mRNA COMM domain containing 1 |
| chr8_-_40634776 | 0.38 |
ENSMUST00000048898.10
ENSMUST00000174205.1 |
Mtmr7
|
myotubularin related protein 7 |
| chr13_-_67755132 | 0.37 |
ENSMUST00000091520.6
|
Zfp85-rs1
|
zinc finger protein 85, related sequence 1 |
| chr1_+_75236439 | 0.37 |
ENSMUST00000082158.6
ENSMUST00000055223.7 |
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr11_+_76904513 | 0.37 |
ENSMUST00000072633.3
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_+_174368919 | 0.37 |
ENSMUST00000085862.1
|
Olfr417
|
olfactory receptor 417 |
| chr3_-_58637201 | 0.34 |
ENSMUST00000041115.6
|
Fam194a
|
family with sequence similarity 194, member A |
| chr16_+_4726357 | 0.34 |
ENSMUST00000154117.1
ENSMUST00000004172.8 |
Hmox2
|
heme oxygenase (decycling) 2 |
| chr6_-_101377342 | 0.33 |
ENSMUST00000151175.1
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr7_+_43791891 | 0.33 |
ENSMUST00000005891.5
|
Klk9
|
kallikrein related-peptidase 9 |
| chr17_-_36958533 | 0.33 |
ENSMUST00000172518.1
|
Znrd1
|
zinc ribbon domain containing, 1 |
| chr2_+_128862947 | 0.32 |
ENSMUST00000110324.1
|
Fbln7
|
fibulin 7 |
| chr8_-_22694061 | 0.31 |
ENSMUST00000131767.1
|
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr11_-_103218421 | 0.31 |
ENSMUST00000103076.1
|
Spata32
|
spermatogenesis associated 32 |
| chr11_+_82764332 | 0.30 |
ENSMUST00000056677.6
|
Zfp830
|
zinc finger protein 830 |
| chr4_-_98823840 | 0.29 |
ENSMUST00000097964.2
|
I0C0044D17Rik
|
RIKEN cDNA I0C0044D17 gene |
| chr2_+_86034765 | 0.29 |
ENSMUST00000062360.4
|
Olfr1033
|
olfactory receptor 1033 |
| chrX_+_37493428 | 0.28 |
ENSMUST00000115179.3
|
Rhox2d
|
reproductive homeobox 2D |
| chr19_+_55742242 | 0.28 |
ENSMUST00000111652.1
ENSMUST00000111649.1 ENSMUST00000111651.1 ENSMUST00000111653.1 ENSMUST00000111656.1 ENSMUST00000127233.1 ENSMUST00000153888.1 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr14_-_78088994 | 0.27 |
ENSMUST00000118785.2
ENSMUST00000066437.4 |
Fam216b
|
family with sequence similarity 216, member B |
| chr15_-_76374926 | 0.27 |
ENSMUST00000071119.6
|
Tssk5
|
testis-specific serine kinase 5 |
| chr7_-_134364859 | 0.26 |
ENSMUST00000172947.1
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr10_+_84576626 | 0.26 |
ENSMUST00000020223.7
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr6_+_125039760 | 0.26 |
ENSMUST00000140131.1
ENSMUST00000032480.7 |
Ing4
|
inhibitor of growth family, member 4 |
| chr14_-_121797670 | 0.26 |
ENSMUST00000100299.3
|
Dock9
|
dedicator of cytokinesis 9 |
| chr12_-_102758672 | 0.25 |
ENSMUST00000174651.1
|
Gm20604
|
predicted gene 20604 |
| chr1_-_163403627 | 0.24 |
ENSMUST00000045138.4
|
Gorab
|
golgin, RAB6-interacting |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 1.1 | 5.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.9 | 2.8 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.8 | 2.4 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.7 | 5.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 2.0 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.6 | 1.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 4.6 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 2.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 | 2.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.5 | 4.7 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.5 | 1.4 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.5 | 1.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.4 | 2.8 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.4 | 1.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.4 | 1.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 10.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 1.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 1.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 3.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 3.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 1.6 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.3 | 1.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 | 1.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 1.2 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 1.9 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 0.6 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.2 | 1.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 2.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 1.5 | GO:1900194 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 5.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.8 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.9 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 3.0 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 | 1.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 1.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 3.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.9 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 7.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 1.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 3.2 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.2 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.7 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 1.1 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.3 | 1.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 4.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 1.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 0.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 3.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 1.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 2.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 3.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 5.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.0 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 6.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0035631 | IkappaB kinase complex(GO:0008385) CD40 receptor complex(GO:0035631) |
| 0.0 | 1.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 4.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.8 | 2.4 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.6 | 1.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.6 | 6.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 2.2 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 4.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.5 | 2.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.5 | 1.4 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.4 | 1.4 | GO:0042903 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.3 | 1.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 1.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 1.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 10.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 4.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 0.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 1.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 1.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 3.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.5 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 2.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:1904315 | GABA-gated chloride ion channel activity(GO:0022851) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 4.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 3.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.1 | 3.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.0 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 3.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 4.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 1.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 8.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 4.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 10.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 5.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |