Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
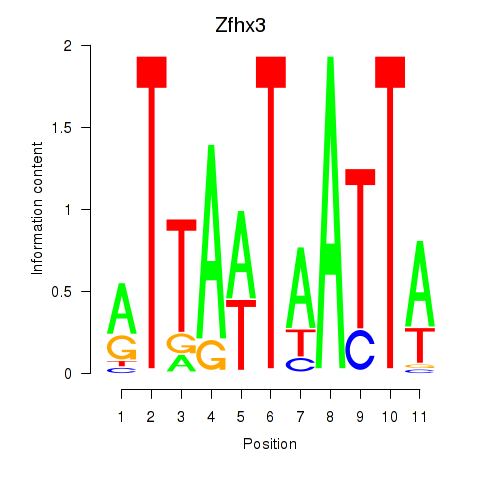
Results for Zfhx3
Z-value: 0.60
Transcription factors associated with Zfhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfhx3
|
ENSMUSG00000038872.8 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | mm10_v2_chr8_+_108714644_108714644 | -0.16 | 3.5e-01 | Click! |
Activity profile of Zfhx3 motif
Sorted Z-values of Zfhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_27000362 | 4.87 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_+_174172738 | 4.62 |
ENSMUST00000027817.7
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr15_-_79285502 | 3.88 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chrX_-_162964557 | 3.66 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr19_+_10015016 | 3.39 |
ENSMUST00000137637.1
ENSMUST00000149967.1 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr1_-_132390301 | 3.21 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr15_-_79285470 | 2.61 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr11_+_24078111 | 2.59 |
ENSMUST00000109516.1
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr11_+_24078022 | 2.43 |
ENSMUST00000000881.6
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr1_-_89933290 | 2.36 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr7_-_4397705 | 2.30 |
ENSMUST00000108590.2
|
Gp6
|
glycoprotein 6 (platelet) |
| chr5_-_43981757 | 2.26 |
ENSMUST00000061299.7
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr3_+_153844209 | 2.24 |
ENSMUST00000044089.3
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr10_+_79927330 | 2.16 |
ENSMUST00000105376.1
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr10_+_79927039 | 2.11 |
ENSMUST00000019708.5
ENSMUST00000105377.1 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_+_24078173 | 2.00 |
ENSMUST00000109514.1
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr6_+_142298419 | 1.85 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr17_-_43502773 | 1.70 |
ENSMUST00000024707.8
ENSMUST00000117137.1 |
Mep1a
|
meprin 1 alpha |
| chrX_+_164140447 | 1.66 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr8_-_107065632 | 1.61 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chrX_-_139871637 | 1.52 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chr1_-_126830632 | 1.45 |
ENSMUST00000112583.1
ENSMUST00000094609.3 |
Nckap5
|
NCK-associated protein 5 |
| chr11_+_3983704 | 1.39 |
ENSMUST00000063004.7
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr7_+_30493622 | 1.33 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr11_+_3983636 | 1.32 |
ENSMUST00000078757.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr19_-_38043559 | 1.31 |
ENSMUST00000041475.8
ENSMUST00000172095.2 |
Myof
|
myoferlin |
| chr18_+_11633276 | 1.31 |
ENSMUST00000115861.2
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr6_-_141773810 | 1.23 |
ENSMUST00000148411.1
|
Gm5724
|
predicted gene 5724 |
| chr2_-_121235689 | 1.20 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr18_+_37518341 | 1.19 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chr11_-_76027726 | 1.18 |
ENSMUST00000021207.6
|
Fam101b
|
family with sequence similarity 101, member B |
| chr4_+_13743424 | 1.16 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_4880669 | 1.06 |
ENSMUST00000078030.3
|
Gm6104
|
predicted gene 6104 |
| chr3_-_17230976 | 1.04 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr2_+_121506748 | 1.00 |
ENSMUST00000099473.3
ENSMUST00000110602.2 |
Wdr76
|
WD repeat domain 76 |
| chr9_-_120068263 | 0.90 |
ENSMUST00000064165.3
ENSMUST00000177637.1 |
Cx3cr1
|
chemokine (C-X3-C) receptor 1 |
| chr3_-_75270073 | 0.89 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr5_+_25246775 | 0.87 |
ENSMUST00000144971.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chr19_-_31765027 | 0.85 |
ENSMUST00000065067.6
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chrX_+_163911401 | 0.85 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_-_30842765 | 0.81 |
ENSMUST00000019924.8
|
Hey2
|
hairy/enhancer-of-split related with YRPW motif 2 |
| chr13_-_58354862 | 0.80 |
ENSMUST00000043605.5
|
Kif27
|
kinesin family member 27 |
| chr13_-_95478655 | 0.80 |
ENSMUST00000022186.3
|
S100z
|
S100 calcium binding protein, zeta |
| chr1_-_66945361 | 0.79 |
ENSMUST00000160100.1
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_-_140671462 | 0.76 |
ENSMUST00000110057.2
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr3_+_136670076 | 0.74 |
ENSMUST00000070198.7
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr17_-_24073479 | 0.73 |
ENSMUST00000017090.5
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr8_+_93810832 | 0.71 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr10_-_44458687 | 0.66 |
ENSMUST00000105490.2
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr12_-_87443800 | 0.66 |
ENSMUST00000162961.1
|
Alkbh1
|
alkB, alkylation repair homolog 1 (E. coli) |
| chr1_-_172895048 | 0.65 |
ENSMUST00000027824.5
|
Apcs
|
serum amyloid P-component |
| chr16_-_74411776 | 0.65 |
ENSMUST00000116586.2
|
Robo2
|
roundabout homolog 2 (Drosophila) |
| chr1_-_138175126 | 0.65 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_79530569 | 0.64 |
ENSMUST00000103236.3
ENSMUST00000170799.1 ENSMUST00000170422.2 |
Evi2a
Evi2b
|
ecotropic viral integration site 2a ecotropic viral integration site 2b |
| chr4_+_150920154 | 0.60 |
ENSMUST00000105672.3
ENSMUST00000030808.3 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr3_-_130730375 | 0.60 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr17_-_50094277 | 0.58 |
ENSMUST00000113195.1
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr4_-_32923455 | 0.56 |
ENSMUST00000035719.4
ENSMUST00000084749.1 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr4_+_101507947 | 0.56 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr2_-_140671400 | 0.54 |
ENSMUST00000056760.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr2_-_165368723 | 0.51 |
ENSMUST00000141140.1
ENSMUST00000103085.1 |
Zfp663
|
zinc finger protein 663 |
| chr6_+_52177498 | 0.51 |
ENSMUST00000070587.3
|
5730596B20Rik
|
RIKEN cDNA 5730596B20 gene |
| chr9_+_95857597 | 0.50 |
ENSMUST00000034980.7
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr8_+_83666827 | 0.49 |
ENSMUST00000019608.5
|
Ptger1
|
prostaglandin E receptor 1 (subtype EP1) |
| chr1_-_138175283 | 0.48 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_122340499 | 0.47 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr3_-_59220150 | 0.46 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr1_-_138175238 | 0.45 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_28263205 | 0.43 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr11_+_4986824 | 0.43 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr15_+_102459193 | 0.39 |
ENSMUST00000164957.1
ENSMUST00000171245.1 |
Prr13
|
proline rich 13 |
| chr10_-_88605017 | 0.38 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr7_+_103928825 | 0.38 |
ENSMUST00000106863.1
|
Olfr631
|
olfactory receptor 631 |
| chr12_+_71170589 | 0.36 |
ENSMUST00000129376.1
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr10_+_23796946 | 0.35 |
ENSMUST00000119597.1
ENSMUST00000179321.1 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr15_+_102459028 | 0.35 |
ENSMUST00000164938.1
ENSMUST00000023810.5 |
Prr13
|
proline rich 13 |
| chr3_+_84952146 | 0.35 |
ENSMUST00000029727.7
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr9_+_53771499 | 0.35 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chrX_+_164373363 | 0.35 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr1_-_126830786 | 0.33 |
ENSMUST00000162646.1
|
Nckap5
|
NCK-associated protein 5 |
| chr10_-_44458715 | 0.33 |
ENSMUST00000039174.4
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chrX_+_112011007 | 0.33 |
ENSMUST00000131304.1
|
Tex16
|
testis expressed gene 16 |
| chr10_-_130394241 | 0.33 |
ENSMUST00000094502.4
|
Vmn2r84
|
vomeronasal 2, receptor 84 |
| chr4_+_82065924 | 0.32 |
ENSMUST00000161588.1
|
Gm5860
|
predicted gene 5860 |
| chr7_+_55794146 | 0.32 |
ENSMUST00000032627.3
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr5_+_107497762 | 0.31 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr4_+_109343029 | 0.31 |
ENSMUST00000030281.5
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr3_-_72967854 | 0.31 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr3_-_69859875 | 0.31 |
ENSMUST00000051239.7
ENSMUST00000171529.1 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr17_-_40880525 | 0.30 |
ENSMUST00000068258.2
|
9130008F23Rik
|
RIKEN cDNA 9130008F23 gene |
| chr11_+_89073841 | 0.28 |
ENSMUST00000100619.4
|
Gm525
|
predicted gene 525 |
| chr2_+_70039114 | 0.28 |
ENSMUST00000060208.4
|
Myo3b
|
myosin IIIB |
| chr3_-_145032765 | 0.28 |
ENSMUST00000029919.5
|
Clca3
|
chloride channel calcium activated 3 |
| chr12_-_54695885 | 0.28 |
ENSMUST00000067272.8
|
Eapp
|
E2F-associated phosphoprotein |
| chr15_+_91231578 | 0.28 |
ENSMUST00000109284.2
|
CN725425
|
cDNA sequence CN725425 |
| chr18_+_55057557 | 0.28 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
| chr13_-_75943812 | 0.28 |
ENSMUST00000022078.5
ENSMUST00000109606.1 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr1_-_136006342 | 0.23 |
ENSMUST00000166193.2
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr9_+_36832684 | 0.23 |
ENSMUST00000034630.8
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr3_+_76075583 | 0.20 |
ENSMUST00000160261.1
|
Fstl5
|
follistatin-like 5 |
| chr12_-_27160498 | 0.19 |
ENSMUST00000182592.1
|
Gm9866
|
predicted gene 9866 |
| chr9_+_113812547 | 0.19 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr5_+_107497718 | 0.17 |
ENSMUST00000112671.2
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr18_+_73863672 | 0.17 |
ENSMUST00000134847.1
|
Mro
|
maestro |
| chr8_-_66486494 | 0.17 |
ENSMUST00000026681.5
|
Tma16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr9_+_85312773 | 0.15 |
ENSMUST00000113205.1
|
Gm11114
|
predicted gene 11114 |
| chr7_+_19119853 | 0.15 |
ENSMUST00000053109.3
|
Fbxo46
|
F-box protein 46 |
| chr12_-_27160311 | 0.14 |
ENSMUST00000182473.1
ENSMUST00000177636.1 ENSMUST00000183238.1 |
Gm9866
|
predicted gene 9866 |
| chr12_-_12941827 | 0.14 |
ENSMUST00000043396.7
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr2_+_78051220 | 0.12 |
ENSMUST00000144728.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr18_+_32815383 | 0.12 |
ENSMUST00000025237.3
|
Tslp
|
thymic stromal lymphopoietin |
| chr1_-_158814469 | 0.11 |
ENSMUST00000161589.2
|
Pappa2
|
pappalysin 2 |
| chr10_+_23797052 | 0.10 |
ENSMUST00000133289.1
|
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr6_-_38124568 | 0.10 |
ENSMUST00000040259.4
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr2_-_72986716 | 0.10 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chrX_+_71972986 | 0.09 |
ENSMUST00000066116.1
|
Fate1
|
fetal and adult testis expressed 1 |
| chrX_+_112240474 | 0.09 |
ENSMUST00000164272.2
ENSMUST00000132037.1 |
4933403O08Rik
|
RIKEN cDNA 4933403O08 gene |
| chr7_-_100583072 | 0.08 |
ENSMUST00000152876.1
ENSMUST00000150042.1 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr6_+_127453667 | 0.08 |
ENSMUST00000112193.1
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr15_-_55548164 | 0.07 |
ENSMUST00000165356.1
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr2_+_88817173 | 0.07 |
ENSMUST00000072057.1
|
Olfr1202
|
olfactory receptor 1202 |
| chr9_-_48964990 | 0.07 |
ENSMUST00000008734.4
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr2_-_140671440 | 0.07 |
ENSMUST00000099301.1
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr16_-_79091078 | 0.06 |
ENSMUST00000023566.4
ENSMUST00000060402.5 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr3_+_125404072 | 0.06 |
ENSMUST00000173932.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr8_+_72107040 | 0.06 |
ENSMUST00000055735.4
|
Olfr374
|
olfactory receptor 374 |
| chr12_-_54695829 | 0.04 |
ENSMUST00000162106.1
ENSMUST00000160085.1 ENSMUST00000161592.1 ENSMUST00000163433.1 |
Eapp
|
E2F-associated phosphoprotein |
| chr5_+_24394388 | 0.04 |
ENSMUST00000115074.1
|
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr1_+_125676969 | 0.04 |
ENSMUST00000027581.6
|
Gpr39
|
G protein-coupled receptor 39 |
| chrX_+_112311334 | 0.03 |
ENSMUST00000026599.3
ENSMUST00000113415.1 |
Apool
|
apolipoprotein O-like |
| chr19_-_31764963 | 0.03 |
ENSMUST00000182685.1
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr5_-_86468990 | 0.03 |
ENSMUST00000101073.2
|
Tmprss11a
|
transmembrane protease, serine 11a |
| chr1_+_179961110 | 0.03 |
ENSMUST00000076687.5
ENSMUST00000097450.3 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_86765807 | 0.02 |
ENSMUST00000123732.1
|
Anxa4
|
annexin A4 |
| chr17_+_35076902 | 0.00 |
ENSMUST00000172494.1
ENSMUST00000172678.1 ENSMUST00000013910.4 |
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr6_-_86765866 | 0.00 |
ENSMUST00000113675.1
|
Anxa4
|
annexin A4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.5 | 4.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 1.4 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.4 | 1.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 1.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 1.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.3 | 1.6 | GO:2000473 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.3 | 2.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 2.4 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.3 | 2.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 | 6.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 0.8 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 | 1.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 0.7 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 1.9 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 0.7 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 0.7 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 0.9 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.2 | 0.5 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 0.7 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 | 0.5 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.8 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 0.6 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 2.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 4.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 1.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 3.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 1.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 6.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 8.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 6.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 3.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 0.8 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 1.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 1.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 2.3 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.2 | 0.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.2 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.7 | GO:0051430 | G-protein coupled serotonin receptor binding(GO:0031821) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.5 | GO:0032407 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 12.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.4 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 5.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 4.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |