Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Arid5a
Z-value: 0.55
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSMUSG00000037447.10 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5a | mm10_v2_chr1_+_36307727_36307735 | -0.76 | 8.7e-08 | Click! |
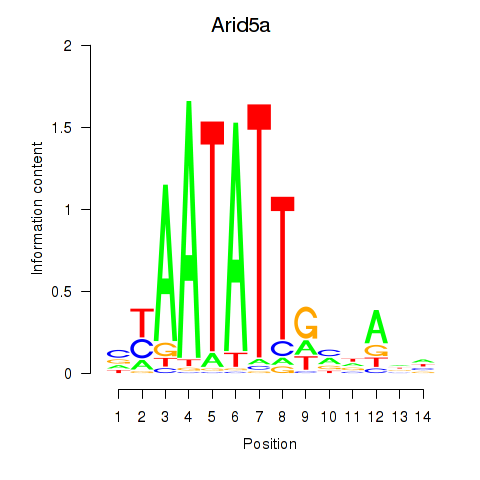
Activity profile of Arid5a motif
Sorted Z-values of Arid5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_58754910 | 2.34 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr2_+_58755177 | 2.33 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr10_-_115362191 | 1.76 |
ENSMUST00000092170.5
|
Tmem19
|
transmembrane protein 19 |
| chr7_-_105600103 | 1.15 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr6_-_119467210 | 1.13 |
ENSMUST00000118120.1
|
Wnt5b
|
wingless-related MMTV integration site 5B |
| chr5_-_31295862 | 0.93 |
ENSMUST00000041266.7
ENSMUST00000172435.1 |
Fndc4
|
fibronectin type III domain containing 4 |
| chrM_+_8600 | 0.37 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr10_+_52391606 | 0.35 |
ENSMUST00000067085.4
|
Nepn
|
nephrocan |
| chr1_-_172057573 | 0.33 |
ENSMUST00000059794.3
|
Nhlh1
|
nescient helix loop helix 1 |
| chr2_+_62046623 | 0.32 |
ENSMUST00000112480.2
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chrM_+_7759 | 0.29 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr2_+_62046580 | 0.25 |
ENSMUST00000054484.8
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr11_-_58534825 | 0.22 |
ENSMUST00000170009.1
|
Olfr330
|
olfactory receptor 330 |
| chrX_-_157415286 | 0.21 |
ENSMUST00000079945.4
ENSMUST00000138396.1 |
Phex
|
phosphate regulating gene with homologies to endopeptidases on the X chromosome (hypophosphatemia, vitamin D resistant rickets) |
| chr2_+_62046462 | 0.16 |
ENSMUST00000102735.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_144033059 | 0.13 |
ENSMUST00000037722.2
ENSMUST00000110032.1 |
Banf2
|
barrier to autointegration factor 2 |
| chr3_-_146781351 | 0.13 |
ENSMUST00000005164.7
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr5_+_68031775 | 0.11 |
ENSMUST00000094715.4
|
Grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr14_+_124005355 | 0.05 |
ENSMUST00000166105.1
|
Gm17615
|
predicted gene, 17615 |
| chr2_+_111780313 | 0.05 |
ENSMUST00000054004.1
|
Olfr1302
|
olfactory receptor 1302 |
| chrX_-_134111852 | 0.01 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr1_-_163085643 | 0.01 |
ENSMUST00000096608.4
|
Mroh9
|
maestro heat-like repeat family member 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 4.6 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 1.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |