Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
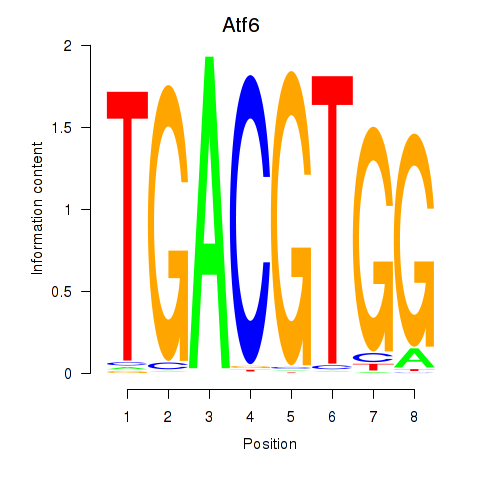
Results for Atf6
Z-value: 0.61
Transcription factors associated with Atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf6
|
ENSMUSG00000026663.6 | activating transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf6 | mm10_v2_chr1_-_170867761_170867784 | -0.15 | 3.7e-01 | Click! |
Activity profile of Atf6 motif
Sorted Z-values of Atf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_36942910 | 1.01 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr7_+_63916857 | 0.93 |
ENSMUST00000177638.1
|
E030018B13Rik
|
RIKEN cDNA E030018B13 gene |
| chr3_+_89459118 | 0.91 |
ENSMUST00000029564.5
|
Pmvk
|
phosphomevalonate kinase |
| chr17_+_36943025 | 0.91 |
ENSMUST00000173072.1
|
Rnf39
|
ring finger protein 39 |
| chr16_-_23988852 | 0.87 |
ENSMUST00000023151.5
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr9_-_86695897 | 0.80 |
ENSMUST00000034989.8
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr11_+_96286623 | 0.78 |
ENSMUST00000049352.7
|
Hoxb7
|
homeobox B7 |
| chr3_+_89459325 | 0.71 |
ENSMUST00000107410.1
|
Pmvk
|
phosphomevalonate kinase |
| chr6_-_124464772 | 0.70 |
ENSMUST00000008297.4
|
Clstn3
|
calsyntenin 3 |
| chr8_+_85492568 | 0.65 |
ENSMUST00000034136.5
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr9_+_44379490 | 0.63 |
ENSMUST00000066601.6
|
Hyou1
|
hypoxia up-regulated 1 |
| chr3_+_123267445 | 0.62 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr11_-_43836243 | 0.59 |
ENSMUST00000167574.1
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr6_-_52246214 | 0.56 |
ENSMUST00000048026.8
|
Hoxa11
|
homeobox A11 |
| chr9_+_44379536 | 0.55 |
ENSMUST00000161318.1
ENSMUST00000160902.1 |
Hyou1
|
hypoxia up-regulated 1 |
| chr2_-_148045891 | 0.54 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr10_-_24101951 | 0.52 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr11_-_69920581 | 0.51 |
ENSMUST00000108610.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr12_-_113260217 | 0.50 |
ENSMUST00000178282.1
|
Igha
|
immunoglobulin heavy constant alpha |
| chr11_+_102393403 | 0.49 |
ENSMUST00000107105.2
ENSMUST00000107102.1 ENSMUST00000107103.1 ENSMUST00000006750.7 |
Rundc3a
|
RUN domain containing 3A |
| chr6_+_83115495 | 0.48 |
ENSMUST00000032114.7
|
Mogs
|
mannosyl-oligosaccharide glucosidase |
| chr9_+_108339048 | 0.47 |
ENSMUST00000082429.5
|
Gpx1
|
glutathione peroxidase 1 |
| chr19_+_8617991 | 0.47 |
ENSMUST00000010250.2
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr8_+_107150621 | 0.45 |
ENSMUST00000034400.3
|
Cyb5b
|
cytochrome b5 type B |
| chrX_+_103356464 | 0.45 |
ENSMUST00000116547.2
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr5_+_9100681 | 0.44 |
ENSMUST00000115365.1
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr18_-_3337614 | 0.44 |
ENSMUST00000150235.1
ENSMUST00000154470.1 |
Crem
|
cAMP responsive element modulator |
| chr1_-_17097839 | 0.44 |
ENSMUST00000038382.4
|
Jph1
|
junctophilin 1 |
| chr11_-_69921329 | 0.43 |
ENSMUST00000108613.3
ENSMUST00000043419.3 ENSMUST00000070996.4 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_+_69991061 | 0.43 |
ENSMUST00000018711.8
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr14_-_26638183 | 0.43 |
ENSMUST00000166902.1
|
4930570N19Rik
|
RIKEN cDNA 4930570N19 gene |
| chr8_-_91801547 | 0.43 |
ENSMUST00000093312.4
|
Irx3
|
Iroquois related homeobox 3 (Drosophila) |
| chr10_-_81291227 | 0.42 |
ENSMUST00000045744.6
|
Tjp3
|
tight junction protein 3 |
| chr11_-_69921190 | 0.41 |
ENSMUST00000108607.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr6_+_134035691 | 0.41 |
ENSMUST00000081028.6
ENSMUST00000111963.1 |
Etv6
|
ets variant gene 6 (TEL oncogene) |
| chr8_-_91801948 | 0.40 |
ENSMUST00000175795.1
|
Irx3
|
Iroquois related homeobox 3 (Drosophila) |
| chr4_-_114908892 | 0.39 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr18_-_3337539 | 0.39 |
ENSMUST00000142690.1
ENSMUST00000025069.4 ENSMUST00000082141.5 ENSMUST00000165086.1 ENSMUST00000149803.1 |
Crem
|
cAMP responsive element modulator |
| chr12_-_57546121 | 0.39 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr18_-_3337467 | 0.38 |
ENSMUST00000154135.1
|
Crem
|
cAMP responsive element modulator |
| chr5_+_124540695 | 0.38 |
ENSMUST00000060226.4
|
Tmed2
|
transmembrane emp24 domain trafficking protein 2 |
| chr2_+_155276297 | 0.38 |
ENSMUST00000029128.3
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr3_-_87795162 | 0.37 |
ENSMUST00000029712.4
|
Ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr7_+_128523576 | 0.37 |
ENSMUST00000033136.7
|
Bag3
|
BCL2-associated athanogene 3 |
| chr4_+_3938888 | 0.37 |
ENSMUST00000121110.1
ENSMUST00000108386.1 ENSMUST00000149544.1 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr11_-_69921057 | 0.35 |
ENSMUST00000108609.1
ENSMUST00000108608.1 ENSMUST00000164359.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_-_136260873 | 0.35 |
ENSMUST00000086395.5
|
Gpr25
|
G protein-coupled receptor 25 |
| chr15_+_7811011 | 0.35 |
ENSMUST00000022744.3
|
Gdnf
|
glial cell line derived neurotrophic factor |
| chr11_-_69920892 | 0.34 |
ENSMUST00000152589.1
ENSMUST00000108612.1 ENSMUST00000108611.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr17_-_37023349 | 0.34 |
ENSMUST00000102665.4
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr14_+_26638237 | 0.34 |
ENSMUST00000112318.3
|
Arf4
|
ADP-ribosylation factor 4 |
| chr5_+_120431770 | 0.34 |
ENSMUST00000031591.7
|
Lhx5
|
LIM homeobox protein 5 |
| chr11_+_96271453 | 0.33 |
ENSMUST00000000010.8
ENSMUST00000174042.1 |
Hoxb9
|
homeobox B9 |
| chr4_+_3938904 | 0.33 |
ENSMUST00000120732.1
ENSMUST00000041122.4 ENSMUST00000121651.1 ENSMUST00000121210.1 ENSMUST00000119307.1 ENSMUST00000123769.1 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr8_-_67818218 | 0.33 |
ENSMUST00000059374.4
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_-_70700070 | 0.32 |
ENSMUST00000116172.1
|
Gm11175
|
predicted gene 11175 |
| chr11_+_84525669 | 0.32 |
ENSMUST00000126072.1
ENSMUST00000128121.1 |
1500016L03Rik
|
RIKEN cDNA 1500016L03 gene |
| chr15_-_98881255 | 0.32 |
ENSMUST00000024518.9
|
Rhebl1
|
Ras homolog enriched in brain like 1 |
| chr1_+_131019843 | 0.32 |
ENSMUST00000016673.5
|
Il10
|
interleukin 10 |
| chr1_-_180245757 | 0.32 |
ENSMUST00000111104.1
|
Psen2
|
presenilin 2 |
| chr2_+_18672384 | 0.31 |
ENSMUST00000171845.1
ENSMUST00000061158.4 |
Commd3
|
COMM domain containing 3 |
| chr11_-_96916448 | 0.30 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr7_-_126898249 | 0.30 |
ENSMUST00000121532.1
ENSMUST00000032926.5 |
Tmem219
|
transmembrane protein 219 |
| chr3_-_30793549 | 0.30 |
ENSMUST00000180833.1
|
4933429H19Rik
|
RIKEN cDNA 4933429H19 gene |
| chr11_+_84525647 | 0.29 |
ENSMUST00000134800.1
|
1500016L03Rik
|
RIKEN cDNA 1500016L03 gene |
| chr6_+_71493850 | 0.29 |
ENSMUST00000064637.4
ENSMUST00000114178.1 |
Rnf103
|
ring finger protein 103 |
| chr5_-_24447587 | 0.28 |
ENSMUST00000127194.1
ENSMUST00000115033.1 ENSMUST00000123167.1 ENSMUST00000030799.8 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr6_+_71494003 | 0.28 |
ENSMUST00000114179.2
|
Rnf103
|
ring finger protein 103 |
| chr7_+_66743504 | 0.28 |
ENSMUST00000066475.8
|
Cers3
|
ceramide synthase 3 |
| chr19_-_5771376 | 0.28 |
ENSMUST00000025890.8
|
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr7_-_80401707 | 0.28 |
ENSMUST00000120753.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr4_+_33031371 | 0.27 |
ENSMUST00000124992.1
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr4_+_47474652 | 0.27 |
ENSMUST00000065678.5
ENSMUST00000137461.1 ENSMUST00000125622.1 |
Sec61b
|
Sec61 beta subunit |
| chr17_-_56183887 | 0.27 |
ENSMUST00000019723.7
|
D17Wsu104e
|
DNA segment, Chr 17, Wayne State University 104, expressed |
| chrX_+_41401128 | 0.27 |
ENSMUST00000115103.2
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr17_-_56074932 | 0.26 |
ENSMUST00000019722.5
|
Ubxn6
|
UBX domain protein 6 |
| chr3_+_30792876 | 0.26 |
ENSMUST00000029256.7
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr11_-_96916366 | 0.26 |
ENSMUST00000144731.1
ENSMUST00000127048.1 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_+_105224314 | 0.26 |
ENSMUST00000068813.2
|
0610012H03Rik
|
RIKEN cDNA 0610012H03 gene |
| chr3_-_144205165 | 0.26 |
ENSMUST00000120539.1
|
Lmo4
|
LIM domain only 4 |
| chr17_+_25875492 | 0.26 |
ENSMUST00000026827.8
ENSMUST00000169308.1 ENSMUST00000169085.1 ENSMUST00000163356.1 |
0610011F06Rik
|
RIKEN cDNA 0610011F06 gene |
| chr2_+_121449362 | 0.26 |
ENSMUST00000110615.1
ENSMUST00000099475.5 |
Serf2
|
small EDRK-rich factor 2 |
| chr4_+_8535604 | 0.25 |
ENSMUST00000060232.7
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr8_-_11550689 | 0.25 |
ENSMUST00000049461.5
|
Cars2
|
cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr3_+_90052814 | 0.25 |
ENSMUST00000160640.1
ENSMUST00000029552.6 ENSMUST00000162114.1 ENSMUST00000068798.6 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr5_+_147860615 | 0.24 |
ENSMUST00000031654.6
|
Pomp
|
proteasome maturation protein |
| chr6_-_31218421 | 0.24 |
ENSMUST00000115107.1
|
AB041803
|
cDNA sequence AB041803 |
| chr4_-_21685782 | 0.24 |
ENSMUST00000076206.4
|
Prdm13
|
PR domain containing 13 |
| chr7_+_127573529 | 0.24 |
ENSMUST00000121004.1
|
Phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr11_-_84525514 | 0.24 |
ENSMUST00000018842.7
|
Lhx1
|
LIM homeobox protein 1 |
| chr19_+_3851797 | 0.23 |
ENSMUST00000072055.6
|
Chka
|
choline kinase alpha |
| chr1_+_171345684 | 0.23 |
ENSMUST00000006579.4
|
Pfdn2
|
prefoldin 2 |
| chr5_-_96161990 | 0.23 |
ENSMUST00000155901.1
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr16_+_20733104 | 0.23 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr5_+_118245226 | 0.23 |
ENSMUST00000049138.7
|
2410131K14Rik
|
RIKEN cDNA 2410131K14 gene |
| chr1_-_131097535 | 0.23 |
ENSMUST00000016672.4
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr2_+_109917639 | 0.23 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr5_+_91139591 | 0.23 |
ENSMUST00000031325.4
|
Areg
|
amphiregulin |
| chr16_-_36874806 | 0.22 |
ENSMUST00000075946.5
|
Eaf2
|
ELL associated factor 2 |
| chr8_-_106136890 | 0.22 |
ENSMUST00000115979.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr8_+_105701624 | 0.22 |
ENSMUST00000093195.6
|
Pard6a
|
par-6 (partitioning defective 6,) homolog alpha (C. elegans) |
| chr8_+_12395287 | 0.22 |
ENSMUST00000180353.1
|
Sox1
|
SRY-box containing gene 1 |
| chr14_+_26638074 | 0.22 |
ENSMUST00000022429.2
|
Arf4
|
ADP-ribosylation factor 4 |
| chr7_-_133776772 | 0.22 |
ENSMUST00000033290.5
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr19_+_3851972 | 0.22 |
ENSMUST00000025760.6
|
Chka
|
choline kinase alpha |
| chr10_-_115315546 | 0.22 |
ENSMUST00000020343.7
|
Rab21
|
RAB21, member RAS oncogene family |
| chr13_-_37994111 | 0.21 |
ENSMUST00000021864.6
|
Ssr1
|
signal sequence receptor, alpha |
| chr13_-_55415166 | 0.21 |
ENSMUST00000054146.3
|
Pfn3
|
profilin 3 |
| chr17_-_56074542 | 0.21 |
ENSMUST00000139371.1
|
Ubxn6
|
UBX domain protein 6 |
| chr7_-_45136391 | 0.21 |
ENSMUST00000146760.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr19_+_3992752 | 0.21 |
ENSMUST00000041871.7
|
Tbx10
|
T-box 10 |
| chr17_-_31636631 | 0.21 |
ENSMUST00000135425.1
ENSMUST00000151718.1 ENSMUST00000155814.1 |
Cbs
|
cystathionine beta-synthase |
| chr14_-_55643251 | 0.20 |
ENSMUST00000120041.1
ENSMUST00000121937.1 ENSMUST00000133707.1 ENSMUST00000002391.8 ENSMUST00000121791.1 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr12_-_69183986 | 0.20 |
ENSMUST00000110620.1
ENSMUST00000110619.1 |
Rpl36al
|
ribosomal protein L36A-like |
| chr7_+_98835104 | 0.20 |
ENSMUST00000165122.1
ENSMUST00000067495.2 |
Wnt11
|
wingless-related MMTV integration site 11 |
| chrX_-_155623325 | 0.20 |
ENSMUST00000038665.5
|
Ptchd1
|
patched domain containing 1 |
| chr7_-_133776681 | 0.20 |
ENSMUST00000130182.1
ENSMUST00000106139.1 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr3_+_107595031 | 0.20 |
ENSMUST00000014747.1
|
Alx3
|
aristaless-like homeobox 3 |
| chr8_+_35375719 | 0.20 |
ENSMUST00000070481.6
|
Ppp1r3b
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B |
| chr2_+_55437100 | 0.20 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr5_+_92137896 | 0.20 |
ENSMUST00000031355.6
|
Uso1
|
USO1 vesicle docking factor |
| chr17_-_34031684 | 0.20 |
ENSMUST00000169397.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_+_120492606 | 0.20 |
ENSMUST00000007139.4
|
Eif1b
|
eukaryotic translation initiation factor 1B |
| chr19_-_47144174 | 0.20 |
ENSMUST00000111813.1
|
Calhm1
|
calcium homeostasis modulator 1 |
| chr12_+_108792946 | 0.19 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chr12_-_69184056 | 0.19 |
ENSMUST00000054544.6
|
Rpl36al
|
ribosomal protein L36A-like |
| chr5_+_28165690 | 0.19 |
ENSMUST00000036177.7
|
En2
|
engrailed 2 |
| chr7_-_45136056 | 0.19 |
ENSMUST00000130628.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr10_+_29211637 | 0.19 |
ENSMUST00000092627.4
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr10_+_80826656 | 0.19 |
ENSMUST00000060987.8
ENSMUST00000177850.1 ENSMUST00000180036.1 ENSMUST00000179172.1 |
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr16_+_17759601 | 0.19 |
ENSMUST00000129199.1
ENSMUST00000165790.1 |
Klhl22
|
kelch-like 22 |
| chr5_-_65492984 | 0.19 |
ENSMUST00000139122.1
|
Smim14
|
small integral membrane protein 14 |
| chr6_+_88084473 | 0.19 |
ENSMUST00000032143.6
|
Rpn1
|
ribophorin I |
| chr8_-_23208407 | 0.19 |
ENSMUST00000167004.1
|
Agpat6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta) |
| chr11_-_115187321 | 0.18 |
ENSMUST00000103038.1
ENSMUST00000103039.1 ENSMUST00000103040.4 |
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_-_91710519 | 0.18 |
ENSMUST00000028678.8
ENSMUST00000076803.5 |
Atg13
|
autophagy related 13 |
| chr4_-_155734627 | 0.18 |
ENSMUST00000178987.1
|
1500002C15Rik
|
RIKEN cDNA 1500002C15 gene |
| chr17_-_36042690 | 0.18 |
ENSMUST00000058801.8
ENSMUST00000080015.5 ENSMUST00000077960.6 |
H2-T22
|
histocompatibility 2, T region locus 22 |
| chr13_-_59557230 | 0.18 |
ENSMUST00000165370.1
ENSMUST00000109830.2 ENSMUST00000022040.6 ENSMUST00000171606.1 ENSMUST00000167096.1 ENSMUST00000166585.1 |
Agtpbp1
|
ATP/GTP binding protein 1 |
| chr8_-_34146974 | 0.18 |
ENSMUST00000033910.8
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr10_+_43901782 | 0.18 |
ENSMUST00000054418.5
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr5_+_143403819 | 0.18 |
ENSMUST00000110731.2
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr4_+_109676568 | 0.18 |
ENSMUST00000102724.4
|
Faf1
|
Fas-associated factor 1 |
| chr10_+_42761483 | 0.18 |
ENSMUST00000019937.4
|
Sec63
|
SEC63-like (S. cerevisiae) |
| chr9_+_59539643 | 0.18 |
ENSMUST00000026262.6
|
Hexa
|
hexosaminidase A |
| chr13_+_73260473 | 0.18 |
ENSMUST00000022095.8
|
Irx4
|
Iroquois related homeobox 4 (Drosophila) |
| chr1_-_161251153 | 0.18 |
ENSMUST00000051925.4
ENSMUST00000071718.5 |
Prdx6
|
peroxiredoxin 6 |
| chr11_-_96916407 | 0.18 |
ENSMUST00000130774.1
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr18_-_84685615 | 0.18 |
ENSMUST00000025546.9
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr4_-_118544010 | 0.18 |
ENSMUST00000128098.1
|
Tmem125
|
transmembrane protein 125 |
| chr5_-_31154152 | 0.18 |
ENSMUST00000114632.1
ENSMUST00000114631.2 ENSMUST00000067186.6 ENSMUST00000137165.1 ENSMUST00000131391.1 ENSMUST00000141823.1 ENSMUST00000154241.1 |
Mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr13_-_100833369 | 0.17 |
ENSMUST00000067246.4
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr4_+_153957230 | 0.17 |
ENSMUST00000058393.2
ENSMUST00000105645.2 |
A430005L14Rik
|
RIKEN cDNA A430005L14 gene |
| chr4_+_125490688 | 0.17 |
ENSMUST00000030676.7
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr12_-_54862783 | 0.17 |
ENSMUST00000078124.7
|
Cfl2
|
cofilin 2, muscle |
| chrX_-_60893430 | 0.17 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr6_+_128644838 | 0.17 |
ENSMUST00000036712.4
|
Gm5884
|
predicted pseudogene 5884 |
| chr5_+_137030275 | 0.17 |
ENSMUST00000041543.8
|
Vgf
|
VGF nerve growth factor inducible |
| chr10_+_120208739 | 0.17 |
ENSMUST00000020446.4
ENSMUST00000134797.1 |
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr5_+_144100387 | 0.17 |
ENSMUST00000041804.7
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr15_+_79030874 | 0.16 |
ENSMUST00000171999.1
ENSMUST00000006544.7 |
Gcat
|
glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
| chr10_+_24076500 | 0.16 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr17_-_65613521 | 0.16 |
ENSMUST00000024897.8
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr17_-_34031544 | 0.16 |
ENSMUST00000025186.8
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr2_-_38926217 | 0.16 |
ENSMUST00000076275.4
ENSMUST00000142130.1 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr13_+_98354234 | 0.16 |
ENSMUST00000105098.3
|
Foxd1
|
forkhead box D1 |
| chr8_-_107056650 | 0.16 |
ENSMUST00000034391.3
ENSMUST00000095517.5 |
Cog8
|
component of oligomeric golgi complex 8 |
| chr16_-_11134624 | 0.16 |
ENSMUST00000038424.7
|
Txndc11
|
thioredoxin domain containing 11 |
| chr11_-_86807624 | 0.16 |
ENSMUST00000018569.7
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr9_+_55208925 | 0.16 |
ENSMUST00000034859.8
|
Fbxo22
|
F-box protein 22 |
| chr2_-_73386396 | 0.16 |
ENSMUST00000112044.1
ENSMUST00000112043.1 ENSMUST00000076463.5 |
Gpr155
|
G protein-coupled receptor 155 |
| chr19_-_41263931 | 0.16 |
ENSMUST00000025989.8
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr13_-_36734450 | 0.16 |
ENSMUST00000037623.8
|
Nrn1
|
neuritin 1 |
| chr8_-_70487314 | 0.16 |
ENSMUST00000045286.7
|
Tmem59l
|
transmembrane protein 59-like |
| chr3_+_122895072 | 0.16 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr11_-_72215592 | 0.15 |
ENSMUST00000021157.8
|
Med31
|
mediator of RNA polymerase II transcription, subunit 31 homolog (yeast) |
| chr19_+_8741413 | 0.15 |
ENSMUST00000176381.1
|
Stx5a
|
syntaxin 5A |
| chr4_+_33031527 | 0.15 |
ENSMUST00000029944.6
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chrX_+_73787062 | 0.15 |
ENSMUST00000002090.2
|
Ssr4
|
signal sequence receptor, delta |
| chr5_+_134986191 | 0.15 |
ENSMUST00000094245.2
|
Cldn3
|
claudin 3 |
| chr6_+_83109071 | 0.15 |
ENSMUST00000113938.3
|
Mrpl53
|
mitochondrial ribosomal protein L53 |
| chr4_+_153957247 | 0.15 |
ENSMUST00000141493.1
|
A430005L14Rik
|
RIKEN cDNA A430005L14 gene |
| chr1_+_58113136 | 0.15 |
ENSMUST00000040999.7
|
Aox3
|
aldehyde oxidase 3 |
| chr7_-_45466894 | 0.15 |
ENSMUST00000033093.8
|
Bax
|
BCL2-associated X protein |
| chr12_+_69184158 | 0.15 |
ENSMUST00000060579.8
|
Mgat2
|
mannoside acetylglucosaminyltransferase 2 |
| chrX_-_136741155 | 0.15 |
ENSMUST00000166930.1
ENSMUST00000113095.1 ENSMUST00000155207.1 ENSMUST00000080411.6 ENSMUST00000169418.1 |
Morf4l2
|
mortality factor 4 like 2 |
| chr11_-_31671863 | 0.15 |
ENSMUST00000058060.7
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr4_-_43000451 | 0.15 |
ENSMUST00000030164.7
|
Vcp
|
valosin containing protein |
| chr2_-_132247623 | 0.15 |
ENSMUST00000110164.1
|
Tmem230
|
transmembrane protein 230 |
| chr5_-_75044562 | 0.15 |
ENSMUST00000075452.5
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr18_+_34331132 | 0.15 |
ENSMUST00000072576.3
ENSMUST00000119329.1 |
Srp19
|
signal recognition particle 19 |
| chr12_-_110978943 | 0.14 |
ENSMUST00000142012.1
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr19_+_21272276 | 0.14 |
ENSMUST00000025659.4
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr8_+_108714644 | 0.14 |
ENSMUST00000043896.8
|
Zfhx3
|
zinc finger homeobox 3 |
| chr4_+_43727181 | 0.14 |
ENSMUST00000095109.3
|
Hrct1
|
histidine rich carboxyl terminus 1 |
| chr7_-_144939823 | 0.14 |
ENSMUST00000093962.4
|
Ccnd1
|
cyclin D1 |
| chr1_-_125912160 | 0.14 |
ENSMUST00000159417.1
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr10_+_125966214 | 0.14 |
ENSMUST00000074807.6
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr10_-_53750880 | 0.14 |
ENSMUST00000020003.7
|
Fam184a
|
family with sequence similarity 184, member A |
| chr2_+_164745979 | 0.14 |
ENSMUST00000017443.7
ENSMUST00000109326.3 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 1.1 | GO:1903382 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.3 | 2.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.9 | GO:0048294 | regulation of memory T cell differentiation(GO:0043380) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 0.6 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 0.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.7 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.8 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 0.3 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.1 | 0.8 | GO:1902031 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.3 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) negative regulation of sensory perception of pain(GO:1904057) |
| 0.1 | 0.2 | GO:0060775 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.9 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.2 | GO:0060067 | cervix development(GO:0060067) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.2 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.6 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.1 | 0.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.6 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 0.2 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:1902445 | B cell negative selection(GO:0002352) B cell homeostatic proliferation(GO:0002358) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.3 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.2 | GO:0060578 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 1.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0070141 | re-entry into mitotic cell cycle(GO:0000320) response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.4 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:1901072 | N-acetylneuraminate catabolic process(GO:0019262) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.5 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 2.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.6 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.5 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.5 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.5 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 1.5 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |