Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
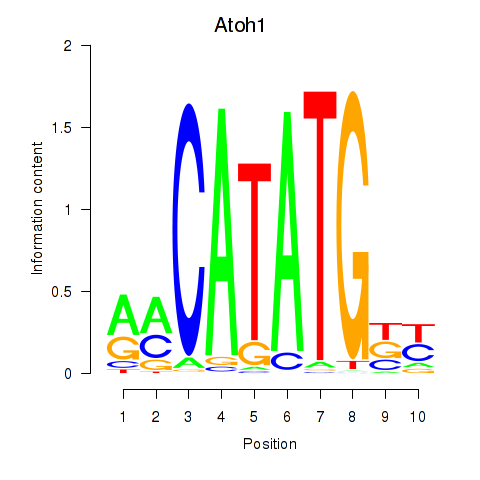
Results for Atoh1_Bhlhe23
Z-value: 0.33
Transcription factors associated with Atoh1_Bhlhe23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atoh1
|
ENSMUSG00000073043.4 | atonal bHLH transcription factor 1 |
|
Bhlhe23
|
ENSMUSG00000045493.3 | basic helix-loop-helix family, member e23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atoh1 | mm10_v2_chr6_+_64729118_64729146 | 0.11 | 5.1e-01 | Click! |
Activity profile of Atoh1_Bhlhe23 motif
Sorted Z-values of Atoh1_Bhlhe23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87861309 | 0.81 |
ENSMUST00000122100.1
|
Igf1
|
insulin-like growth factor 1 |
| chr6_-_87690819 | 0.67 |
ENSMUST00000162547.1
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene |
| chr7_+_30458280 | 0.63 |
ENSMUST00000126297.1
|
Nphs1
|
nephrosis 1, nephrin |
| chr3_+_3508024 | 0.40 |
ENSMUST00000108393.1
ENSMUST00000017832.8 |
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr16_+_43235856 | 0.39 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_+_124194894 | 0.35 |
ENSMUST00000159053.1
ENSMUST00000162577.1 |
Gm16338
|
predicted gene 16338 |
| chr10_-_25200110 | 0.30 |
ENSMUST00000100012.2
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr17_+_36898110 | 0.29 |
ENSMUST00000078438.4
|
Trim31
|
tripartite motif-containing 31 |
| chr1_+_167618246 | 0.28 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr9_+_66946057 | 0.27 |
ENSMUST00000040917.7
ENSMUST00000127896.1 |
Rps27l
|
ribosomal protein S27-like |
| chr2_+_118772766 | 0.27 |
ENSMUST00000130293.1
ENSMUST00000061360.3 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr3_-_84259812 | 0.25 |
ENSMUST00000107691.1
|
Trim2
|
tripartite motif-containing 2 |
| chrX_+_103321398 | 0.24 |
ENSMUST00000033689.2
|
Cdx4
|
caudal type homeobox 4 |
| chr4_+_86053887 | 0.24 |
ENSMUST00000107178.2
ENSMUST00000048885.5 ENSMUST00000141889.1 ENSMUST00000120678.1 |
Adamtsl1
|
ADAMTS-like 1 |
| chr5_-_66618772 | 0.22 |
ENSMUST00000162994.1
ENSMUST00000159512.1 ENSMUST00000159786.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr18_+_34759551 | 0.21 |
ENSMUST00000097622.3
|
Fam53c
|
family with sequence similarity 53, member C |
| chr4_-_82850721 | 0.21 |
ENSMUST00000139401.1
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr4_+_11579647 | 0.20 |
ENSMUST00000180239.1
|
Fsbp
|
fibrinogen silencer binding protein |
| chr11_-_69920581 | 0.20 |
ENSMUST00000108610.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_-_53773187 | 0.19 |
ENSMUST00000170390.1
|
Gm17334
|
predicted gene, 17334 |
| chr11_+_78503449 | 0.19 |
ENSMUST00000001130.6
ENSMUST00000125670.2 |
Sebox
|
SEBOX homeobox |
| chr11_+_117232254 | 0.19 |
ENSMUST00000106354.2
|
Sept9
|
septin 9 |
| chr9_+_24283433 | 0.18 |
ENSMUST00000154644.1
|
Npsr1
|
neuropeptide S receptor 1 |
| chr17_+_82539258 | 0.18 |
ENSMUST00000097278.3
|
Gm6594
|
predicted pseudogene 6594 |
| chr11_-_87404380 | 0.17 |
ENSMUST00000067692.6
|
Rad51c
|
RAD51 homolog C |
| chr19_-_37176055 | 0.17 |
ENSMUST00000142973.1
ENSMUST00000154376.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_-_79456750 | 0.16 |
ENSMUST00000041099.4
|
Neurod1
|
neurogenic differentiation 1 |
| chrX_+_36328353 | 0.16 |
ENSMUST00000016383.3
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr2_+_137663424 | 0.15 |
ENSMUST00000134833.1
|
Gm14064
|
predicted gene 14064 |
| chr11_-_96747419 | 0.15 |
ENSMUST00000181758.1
|
2010300F17Rik
|
RIKEN cDNA 2010300F17 gene |
| chr3_+_118430299 | 0.13 |
ENSMUST00000180774.1
|
Gm26871
|
predicted gene, 26871 |
| chr14_+_28511344 | 0.13 |
ENSMUST00000112272.1
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr6_+_147032528 | 0.12 |
ENSMUST00000036194.4
|
Rep15
|
RAB15 effector protein |
| chr9_-_99717259 | 0.12 |
ENSMUST00000112882.2
ENSMUST00000131922.1 |
Cldn18
|
claudin 18 |
| chr11_-_101417615 | 0.12 |
ENSMUST00000070395.8
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr7_+_17972124 | 0.12 |
ENSMUST00000094799.2
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr7_+_18065929 | 0.12 |
ENSMUST00000032520.2
ENSMUST00000108487.2 ENSMUST00000108483.1 |
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr2_-_52558539 | 0.11 |
ENSMUST00000102760.3
ENSMUST00000102761.2 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr15_-_81408261 | 0.11 |
ENSMUST00000057236.3
|
Dnajb7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr2_+_62046623 | 0.11 |
ENSMUST00000112480.2
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_62046580 | 0.11 |
ENSMUST00000054484.8
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr16_+_43510267 | 0.11 |
ENSMUST00000114695.2
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_-_90129339 | 0.11 |
ENSMUST00000181189.1
|
2310010J17Rik
|
RIKEN cDNA 2310010J17 gene |
| chr13_+_108046411 | 0.10 |
ENSMUST00000095458.4
|
Smim15
|
small integral membrane protein 15 |
| chr8_+_94838321 | 0.10 |
ENSMUST00000034234.8
ENSMUST00000159871.1 |
Coq9
|
coenzyme Q9 homolog (yeast) |
| chr1_+_15287259 | 0.10 |
ENSMUST00000175681.1
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr17_-_25792284 | 0.09 |
ENSMUST00000072735.7
|
Fam173a
|
family with sequence similarity 173, member A |
| chr8_+_25601591 | 0.09 |
ENSMUST00000155861.1
|
Whsc1l1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 (human) |
| chr2_+_127587214 | 0.09 |
ENSMUST00000028852.6
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr11_+_53433299 | 0.09 |
ENSMUST00000018382.6
|
Gdf9
|
growth differentiation factor 9 |
| chr19_-_53589067 | 0.09 |
ENSMUST00000095978.3
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr11_-_98329641 | 0.09 |
ENSMUST00000041685.6
|
Neurod2
|
neurogenic differentiation 2 |
| chr8_+_25602236 | 0.09 |
ENSMUST00000146919.1
ENSMUST00000142395.1 ENSMUST00000139966.1 |
Whsc1l1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 (human) |
| chr2_-_105399286 | 0.09 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr16_+_11008898 | 0.08 |
ENSMUST00000180624.1
|
Gm4262
|
predicted gene 4262 |
| chr13_-_97747399 | 0.08 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr7_+_101394361 | 0.08 |
ENSMUST00000154239.1
ENSMUST00000098243.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr9_+_26999668 | 0.08 |
ENSMUST00000039161.8
|
Thyn1
|
thymocyte nuclear protein 1 |
| chr5_-_66618636 | 0.08 |
ENSMUST00000162382.1
ENSMUST00000160870.1 ENSMUST00000087256.5 ENSMUST00000160103.1 ENSMUST00000162349.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_+_108920342 | 0.08 |
ENSMUST00000052915.7
|
Axin2
|
axin2 |
| chr14_-_16249675 | 0.08 |
ENSMUST00000022311.4
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr3_-_129804030 | 0.08 |
ENSMUST00000179187.1
|
Lrit3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr11_-_35980473 | 0.08 |
ENSMUST00000018993.6
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr4_-_25281801 | 0.07 |
ENSMUST00000102994.3
|
Ufl1
|
UFM1 specific ligase 1 |
| chr12_+_86734381 | 0.07 |
ENSMUST00000095527.5
|
Gm6772
|
predicted gene 6772 |
| chr2_+_62046462 | 0.07 |
ENSMUST00000102735.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr9_+_72985568 | 0.07 |
ENSMUST00000150826.2
ENSMUST00000085350.4 ENSMUST00000140675.1 |
Ccpg1
|
cell cycle progression 1 |
| chr17_-_47016956 | 0.07 |
ENSMUST00000165525.1
|
Gm16494
|
predicted gene 16494 |
| chr6_-_128275577 | 0.07 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr2_+_74727074 | 0.07 |
ENSMUST00000111980.2
|
Hoxd3
|
homeobox D3 |
| chr2_-_54085542 | 0.07 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr10_-_22149270 | 0.07 |
ENSMUST00000179054.1
ENSMUST00000069372.6 |
E030030I06Rik
|
RIKEN cDNA E030030I06 gene |
| chr7_-_141437829 | 0.06 |
ENSMUST00000019226.7
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr15_-_58364148 | 0.06 |
ENSMUST00000068515.7
|
Anxa13
|
annexin A13 |
| chr11_-_94704499 | 0.06 |
ENSMUST00000069852.1
|
Gm11541
|
predicted gene 11541 |
| chr9_+_78175898 | 0.06 |
ENSMUST00000180974.1
|
C920006O11Rik
|
RIKEN cDNA C920006O11 gene |
| chr15_+_78913916 | 0.06 |
ENSMUST00000089378.4
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr1_+_177444653 | 0.06 |
ENSMUST00000094276.3
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr6_-_113501818 | 0.06 |
ENSMUST00000101059.1
|
Prrt3
|
proline-rich transmembrane protein 3 |
| chr16_+_95258036 | 0.06 |
ENSMUST00000113859.1
|
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_-_141437587 | 0.06 |
ENSMUST00000172654.1
ENSMUST00000106006.1 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr5_-_5479120 | 0.05 |
ENSMUST00000115447.1
|
1700015F17Rik
|
RIKEN cDNA 1700015F17 gene |
| chr3_-_123236134 | 0.05 |
ENSMUST00000106427.1
ENSMUST00000106426.1 ENSMUST00000051443.5 |
Synpo2
|
synaptopodin 2 |
| chr6_+_17743582 | 0.05 |
ENSMUST00000000674.6
ENSMUST00000077080.2 |
St7
|
suppression of tumorigenicity 7 |
| chr1_+_177445660 | 0.05 |
ENSMUST00000077225.6
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_-_66618752 | 0.05 |
ENSMUST00000162366.1
|
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_+_60479915 | 0.05 |
ENSMUST00000126522.1
|
Myo15
|
myosin XV |
| chr18_+_55057557 | 0.05 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
| chr14_-_79223876 | 0.05 |
ENSMUST00000040802.4
|
Zfp957
|
zinc finger protein 957 |
| chr1_+_6730051 | 0.05 |
ENSMUST00000043578.6
ENSMUST00000131467.1 ENSMUST00000150761.1 ENSMUST00000151281.1 |
St18
|
suppression of tumorigenicity 18 |
| chr6_+_83034173 | 0.05 |
ENSMUST00000000707.2
ENSMUST00000101257.3 |
Loxl3
|
lysyl oxidase-like 3 |
| chr10_+_61175206 | 0.05 |
ENSMUST00000079235.5
|
Tbata
|
thymus, brain and testes associated |
| chr9_-_26999491 | 0.05 |
ENSMUST00000060513.7
ENSMUST00000120367.1 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chr1_+_6730135 | 0.05 |
ENSMUST00000155921.1
|
St18
|
suppression of tumorigenicity 18 |
| chr1_-_178337774 | 0.04 |
ENSMUST00000037748.7
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U |
| chr7_-_133602110 | 0.04 |
ENSMUST00000033275.2
|
Tex36
|
testis expressed 36 |
| chr19_-_11856001 | 0.04 |
ENSMUST00000079875.3
|
Olfr1418
|
olfactory receptor 1418 |
| chr10_-_92164666 | 0.04 |
ENSMUST00000183123.1
ENSMUST00000182033.1 |
Rmst
|
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
| chr5_+_113226909 | 0.04 |
ENSMUST00000086615.2
|
Tmem211
|
transmembrane protein 211 |
| chr16_+_57353271 | 0.04 |
ENSMUST00000099667.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_+_53596936 | 0.04 |
ENSMUST00000020004.6
|
Asf1a
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae) |
| chr16_+_95258209 | 0.04 |
ENSMUST00000113858.1
|
Kcnj15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_+_100320596 | 0.04 |
ENSMUST00000152521.1
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chrY_-_30840278 | 0.04 |
ENSMUST00000180175.1
|
Gm21529
|
predicted gene, 21529 |
| chr10_+_80142295 | 0.03 |
ENSMUST00000003156.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr2_-_39065505 | 0.03 |
ENSMUST00000039165.8
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr15_+_84669565 | 0.03 |
ENSMUST00000171460.1
|
Prr5
|
proline rich 5 (renal) |
| chr5_-_28210168 | 0.03 |
ENSMUST00000117098.1
|
Cnpy1
|
canopy 1 homolog (zebrafish) |
| chr1_-_183147461 | 0.03 |
ENSMUST00000171366.1
|
Disp1
|
dispatched homolog 1 (Drosophila) |
| chr17_-_6449571 | 0.03 |
ENSMUST00000180035.1
|
Tmem181b-ps
|
transmembrane protein 181B, pseudogene |
| chrX_-_160735747 | 0.03 |
ENSMUST00000135856.1
|
Ppef1
|
protein phosphatase with EF hand calcium-binding domain 1 |
| chr2_+_21367532 | 0.03 |
ENSMUST00000055946.7
|
Gpr158
|
G protein-coupled receptor 158 |
| chr6_-_148831395 | 0.03 |
ENSMUST00000145960.1
|
Ipo8
|
importin 8 |
| chr2_+_78051155 | 0.03 |
ENSMUST00000145972.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr10_+_80142358 | 0.03 |
ENSMUST00000105366.1
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr19_-_37207293 | 0.03 |
ENSMUST00000132580.1
ENSMUST00000079754.4 ENSMUST00000136286.1 ENSMUST00000126188.1 ENSMUST00000126781.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr3_+_125404072 | 0.03 |
ENSMUST00000173932.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr1_+_150392794 | 0.03 |
ENSMUST00000124973.2
|
Tpr
|
translocated promoter region |
| chr9_+_35669624 | 0.03 |
ENSMUST00000118254.1
|
Pate2
|
prostate and testis expressed 2 |
| chr5_-_24447587 | 0.03 |
ENSMUST00000127194.1
ENSMUST00000115033.1 ENSMUST00000123167.1 ENSMUST00000030799.8 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr17_-_6827990 | 0.03 |
ENSMUST00000181895.1
|
Gm2885
|
predicted gene 2885 |
| chr9_-_88719798 | 0.03 |
ENSMUST00000113110.3
|
Gm2382
|
predicted gene 2382 |
| chr14_-_51071442 | 0.02 |
ENSMUST00000048478.5
|
Olfr750
|
olfactory receptor 750 |
| chr5_-_74068361 | 0.02 |
ENSMUST00000119154.1
ENSMUST00000068058.7 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr4_+_127172866 | 0.02 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr2_-_39065438 | 0.02 |
ENSMUST00000112850.2
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr17_+_35552128 | 0.02 |
ENSMUST00000044804.7
|
Cdsn
|
corneodesmosin |
| chr16_+_16213318 | 0.02 |
ENSMUST00000162150.1
ENSMUST00000161342.1 ENSMUST00000039408.2 |
Pkp2
|
plakophilin 2 |
| chr16_-_57292845 | 0.02 |
ENSMUST00000023434.8
ENSMUST00000120112.1 ENSMUST00000119407.1 |
Tmem30c
|
transmembrane protein 30C |
| chr10_+_69706326 | 0.02 |
ENSMUST00000182992.1
|
Ank3
|
ankyrin 3, epithelial |
| chrY_-_3410167 | 0.02 |
ENSMUST00000169382.2
|
Gm21704
|
predicted gene, 21704 |
| chr11_+_67774608 | 0.02 |
ENSMUST00000181566.1
|
Gm12302
|
predicted gene 12302 |
| chr2_-_156180135 | 0.02 |
ENSMUST00000126992.1
ENSMUST00000146288.1 ENSMUST00000029149.6 ENSMUST00000109587.2 ENSMUST00000109584.1 |
Rbm39
|
RNA binding motif protein 39 |
| chr19_-_53371766 | 0.02 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr1_-_150392719 | 0.02 |
ENSMUST00000006167.6
ENSMUST00000094477.2 ENSMUST00000097547.3 |
BC003331
|
cDNA sequence BC003331 |
| chr17_+_86917348 | 0.02 |
ENSMUST00000042172.5
|
Tmem247
|
transmembrane protein 247 |
| chr17_+_35126316 | 0.02 |
ENSMUST00000061859.6
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr15_+_30172570 | 0.02 |
ENSMUST00000081728.5
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr4_+_55350043 | 0.02 |
ENSMUST00000030134.8
|
Rad23b
|
RAD23b homolog (S. cerevisiae) |
| chr6_-_3494587 | 0.01 |
ENSMUST00000049985.8
|
Hepacam2
|
HEPACAM family member 2 |
| chr17_-_86947867 | 0.01 |
ENSMUST00000065758.7
|
Atp6v1e2
|
ATPase, H+ transporting, lysosomal V1 subunit E2 |
| chr13_+_37345338 | 0.01 |
ENSMUST00000021860.5
|
Ly86
|
lymphocyte antigen 86 |
| chr2_+_164833841 | 0.01 |
ENSMUST00000152721.1
|
Ctsa
|
cathepsin A |
| chr4_-_141598206 | 0.01 |
ENSMUST00000131317.1
ENSMUST00000006381.4 ENSMUST00000129602.1 |
Fblim1
|
filamin binding LIM protein 1 |
| chr1_-_111864869 | 0.01 |
ENSMUST00000035462.5
|
Dsel
|
dermatan sulfate epimerase-like |
| chr11_+_49247462 | 0.01 |
ENSMUST00000109194.1
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr11_+_31872100 | 0.01 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr18_-_77186257 | 0.01 |
ENSMUST00000097520.2
|
Gm7276
|
predicted gene 7276 |
| chr2_+_69670100 | 0.01 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr5_+_3343893 | 0.00 |
ENSMUST00000165117.1
|
Cdk6
|
cyclin-dependent kinase 6 |
| chr15_+_25622525 | 0.00 |
ENSMUST00000110457.1
ENSMUST00000137601.1 |
Myo10
|
myosin X |
| chr8_+_4243264 | 0.00 |
ENSMUST00000110996.1
|
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr2_-_80128834 | 0.00 |
ENSMUST00000102654.4
ENSMUST00000102655.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atoh1_Bhlhe23
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.2 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.2 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:0061349 | cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.0 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.8 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |