Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Crem_Jdp2
Z-value: 1.01
Transcription factors associated with Crem_Jdp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
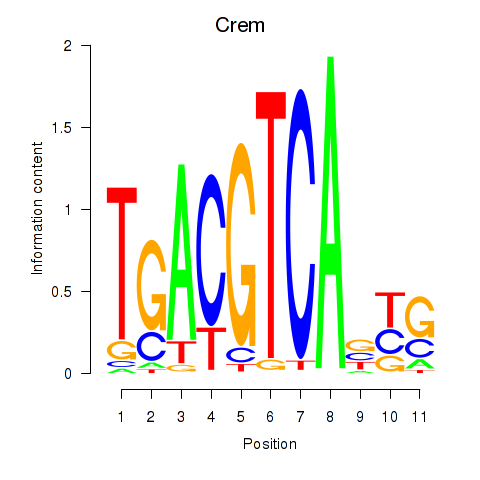
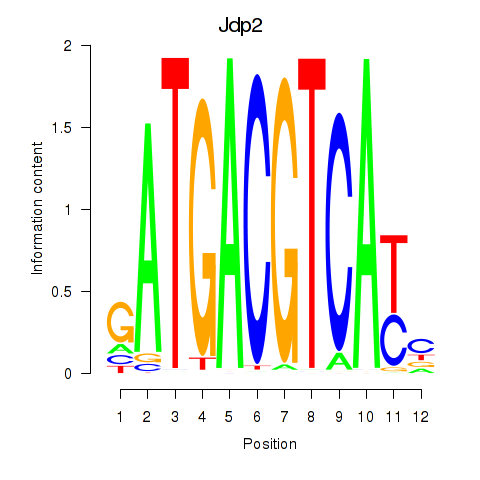
Crem
|
ENSMUSG00000063889.10 | cAMP responsive element modulator |
|
Jdp2
|
ENSMUSG00000034271.9 | Jun dimerization protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Crem | mm10_v2_chr18_-_3281036_3281132 | 0.67 | 7.4e-06 | Click! |
| Jdp2 | mm10_v2_chr12_+_85599047_85599105 | -0.23 | 1.8e-01 | Click! |
Activity profile of Crem_Jdp2 motif
Sorted Z-values of Crem_Jdp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_155276297 | 4.83 |
ENSMUST00000029128.3
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr18_-_3281036 | 3.60 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr18_-_3281712 | 2.81 |
ENSMUST00000182204.1
ENSMUST00000154705.1 ENSMUST00000182833.1 ENSMUST00000151084.1 |
Crem
|
cAMP responsive element modulator |
| chr3_-_20275659 | 2.79 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr1_+_71652837 | 2.76 |
ENSMUST00000097699.2
|
Apol7d
|
apolipoprotein L 7d |
| chr2_+_143546144 | 2.70 |
ENSMUST00000028905.9
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr1_-_79440039 | 2.62 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chr18_-_3281752 | 2.61 |
ENSMUST00000140332.1
ENSMUST00000147138.1 |
Crem
|
cAMP responsive element modulator |
| chr4_+_141301228 | 2.04 |
ENSMUST00000006614.2
|
Epha2
|
Eph receptor A2 |
| chr16_-_23890805 | 1.97 |
ENSMUST00000004480.3
|
Sst
|
somatostatin |
| chr10_-_115587739 | 1.91 |
ENSMUST00000020350.8
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr7_+_112679327 | 1.89 |
ENSMUST00000106638.2
|
Tead1
|
TEA domain family member 1 |
| chr2_+_173153048 | 1.87 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr15_+_25843264 | 1.86 |
ENSMUST00000022881.7
|
Fam134b
|
family with sequence similarity 134, member B |
| chr10_+_44268328 | 1.84 |
ENSMUST00000039286.4
|
Atg5
|
autophagy related 5 |
| chr1_-_71653162 | 1.79 |
ENSMUST00000055226.6
|
Fn1
|
fibronectin 1 |
| chr9_-_96752822 | 1.71 |
ENSMUST00000152594.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr12_-_86884808 | 1.70 |
ENSMUST00000038422.6
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr4_+_11704439 | 1.68 |
ENSMUST00000108304.2
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr17_+_43953191 | 1.62 |
ENSMUST00000044792.4
|
Rcan2
|
regulator of calcineurin 2 |
| chr2_+_155382186 | 1.52 |
ENSMUST00000134218.1
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr16_-_4790220 | 1.50 |
ENSMUST00000118703.1
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr17_-_32420965 | 1.48 |
ENSMUST00000170392.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr14_+_70577839 | 1.44 |
ENSMUST00000089049.2
|
Nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr9_+_77917364 | 1.37 |
ENSMUST00000034904.7
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr6_+_129533183 | 1.36 |
ENSMUST00000032264.6
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_-_19310035 | 1.33 |
ENSMUST00000003640.2
|
Fosb
|
FBJ osteosarcoma oncogene B |
| chr19_+_23141183 | 1.32 |
ENSMUST00000036884.1
|
Klf9
|
Kruppel-like factor 9 |
| chr17_+_43952999 | 1.32 |
ENSMUST00000177857.1
|
Rcan2
|
regulator of calcineurin 2 |
| chr16_-_4789887 | 1.28 |
ENSMUST00000117713.1
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr3_-_88548249 | 1.27 |
ENSMUST00000131775.1
ENSMUST00000008745.6 |
Rab25
|
RAB25, member RAS oncogene family |
| chr6_-_113531575 | 1.24 |
ENSMUST00000032425.5
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr9_-_96752653 | 1.23 |
ENSMUST00000062899.3
|
E030011O05Rik
|
RIKEN cDNA E030011O05 gene |
| chr8_-_24438937 | 1.22 |
ENSMUST00000052622.4
|
1810011O10Rik
|
RIKEN cDNA 1810011O10 gene |
| chr8_-_3467617 | 1.21 |
ENSMUST00000111081.3
ENSMUST00000118194.1 ENSMUST00000004686.6 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr3_-_85741389 | 1.20 |
ENSMUST00000094148.4
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr13_-_54688184 | 1.19 |
ENSMUST00000150806.1
ENSMUST00000125927.1 |
Rnf44
|
ring finger protein 44 |
| chr2_-_148045891 | 1.18 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr15_-_68363139 | 1.18 |
ENSMUST00000175699.1
|
Gm20732
|
predicted gene 20732 |
| chr17_-_26069409 | 1.18 |
ENSMUST00000120691.1
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr11_+_101665541 | 1.17 |
ENSMUST00000039388.2
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chr1_-_36709904 | 1.16 |
ENSMUST00000043951.3
|
Actr1b
|
ARP1 actin-related protein 1B, centractin beta |
| chr14_-_59597836 | 1.16 |
ENSMUST00000167100.1
ENSMUST00000022555.4 ENSMUST00000056997.7 ENSMUST00000171683.1 |
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chrX_-_20920911 | 1.15 |
ENSMUST00000081893.6
ENSMUST00000115345.1 |
Syn1
|
synapsin I |
| chr7_+_125707945 | 1.15 |
ENSMUST00000148701.1
|
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr16_-_4789984 | 1.15 |
ENSMUST00000004173.5
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr17_-_46487641 | 1.14 |
ENSMUST00000047034.8
|
Ttbk1
|
tau tubulin kinase 1 |
| chr2_-_32424005 | 1.08 |
ENSMUST00000113307.2
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr17_-_35979679 | 1.07 |
ENSMUST00000173724.1
ENSMUST00000172900.1 ENSMUST00000174849.1 |
Prr3
|
proline-rich polypeptide 3 |
| chr7_-_138846202 | 1.02 |
ENSMUST00000118810.1
ENSMUST00000075667.4 ENSMUST00000119664.1 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr10_+_80141457 | 1.00 |
ENSMUST00000105367.1
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr2_-_130642770 | 0.99 |
ENSMUST00000045761.6
|
Lzts3
|
leucine zipper, putative tumor suppressor family member 3 |
| chr9_+_44066993 | 0.99 |
ENSMUST00000034508.7
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr9_+_44067072 | 0.98 |
ENSMUST00000177054.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr1_+_24678536 | 0.98 |
ENSMUST00000095062.3
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr5_+_91139591 | 0.96 |
ENSMUST00000031325.4
|
Areg
|
amphiregulin |
| chr9_+_46012810 | 0.96 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr17_-_33760306 | 0.96 |
ENSMUST00000173860.1
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr9_-_52679429 | 0.95 |
ENSMUST00000098768.2
|
AI593442
|
expressed sequence AI593442 |
| chr16_-_24393588 | 0.95 |
ENSMUST00000181640.1
|
1110054M08Rik
|
RIKEN cDNA 1110054M08 gene |
| chr13_-_64274962 | 0.94 |
ENSMUST00000039318.8
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr8_+_123373778 | 0.94 |
ENSMUST00000057934.3
ENSMUST00000108840.2 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr10_-_20724696 | 0.94 |
ENSMUST00000170265.1
|
Pde7b
|
phosphodiesterase 7B |
| chr13_-_64274879 | 0.94 |
ENSMUST00000109770.1
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr7_+_112679314 | 0.91 |
ENSMUST00000084705.5
ENSMUST00000059768.10 |
Tead1
|
TEA domain family member 1 |
| chr11_-_119547744 | 0.91 |
ENSMUST00000026670.4
|
Nptx1
|
neuronal pentraxin 1 |
| chr7_+_102210335 | 0.91 |
ENSMUST00000140631.1
ENSMUST00000120879.1 ENSMUST00000146996.1 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr9_+_102718424 | 0.90 |
ENSMUST00000156485.1
ENSMUST00000145937.1 ENSMUST00000134483.1 |
Amotl2
|
angiomotin-like 2 |
| chr11_+_79660532 | 0.90 |
ENSMUST00000155381.1
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr6_-_85933379 | 0.90 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr5_-_124032214 | 0.89 |
ENSMUST00000040967.7
|
Vps37b
|
vacuolar protein sorting 37B (yeast) |
| chr6_-_115592571 | 0.89 |
ENSMUST00000112957.1
|
2510049J12Rik
|
RIKEN cDNA 2510049J12 gene |
| chr15_+_99393219 | 0.86 |
ENSMUST00000159209.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr11_+_101468164 | 0.86 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr10_-_20725023 | 0.86 |
ENSMUST00000020165.7
|
Pde7b
|
phosphodiesterase 7B |
| chr9_+_59578192 | 0.86 |
ENSMUST00000118549.1
ENSMUST00000034840.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr7_+_125707893 | 0.86 |
ENSMUST00000069660.6
ENSMUST00000142464.1 |
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr8_+_11728105 | 0.85 |
ENSMUST00000110909.2
ENSMUST00000033908.6 |
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr15_+_99393574 | 0.85 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_+_92741603 | 0.85 |
ENSMUST00000032879.7
|
Rab30
|
RAB30, member RAS oncogene family |
| chr3_-_138143352 | 0.83 |
ENSMUST00000098580.2
|
Mttp
|
microsomal triglyceride transfer protein |
| chr15_+_99393610 | 0.82 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_-_184033998 | 0.82 |
ENSMUST00000050306.5
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr12_+_108792946 | 0.81 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chr15_+_34238026 | 0.81 |
ENSMUST00000022867.3
|
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr4_-_109202217 | 0.80 |
ENSMUST00000160774.1
ENSMUST00000030288.7 ENSMUST00000162787.2 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr9_+_46012822 | 0.80 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr3_-_88762244 | 0.79 |
ENSMUST00000183267.1
|
Syt11
|
synaptotagmin XI |
| chr17_+_33432890 | 0.78 |
ENSMUST00000174088.2
|
Actl9
|
actin-like 9 |
| chr16_+_56477838 | 0.78 |
ENSMUST00000048471.7
ENSMUST00000096013.3 ENSMUST00000096012.3 ENSMUST00000171000.1 |
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein |
| chr8_+_123235086 | 0.77 |
ENSMUST00000181432.1
|
4933417D19Rik
|
RIKEN cDNA 4933417D19 gene |
| chr16_-_97962581 | 0.76 |
ENSMUST00000113734.2
ENSMUST00000052089.7 ENSMUST00000063605.7 |
Zbtb21
|
zinc finger and BTB domain containing 21 |
| chr10_-_102490418 | 0.76 |
ENSMUST00000020040.3
|
Nts
|
neurotensin |
| chr1_-_161034794 | 0.76 |
ENSMUST00000177003.1
ENSMUST00000162226.2 ENSMUST00000159250.2 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr1_+_74791516 | 0.75 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr16_-_16829276 | 0.75 |
ENSMUST00000023468.5
|
Spag6
|
sperm associated antigen 6 |
| chr2_+_30237680 | 0.74 |
ENSMUST00000113654.1
ENSMUST00000095078.2 |
Lrrc8a
|
leucine rich repeat containing 8A |
| chr14_-_73385225 | 0.74 |
ENSMUST00000022704.7
|
Itm2b
|
integral membrane protein 2B |
| chr2_+_112454997 | 0.74 |
ENSMUST00000069747.5
|
Emc7
|
ER membrane protein complex subunit 7 |
| chr11_+_52396414 | 0.73 |
ENSMUST00000109057.1
ENSMUST00000036952.4 |
9530068E07Rik
|
RIKEN cDNA 9530068E07 gene |
| chr18_+_36348739 | 0.71 |
ENSMUST00000152804.2
|
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr5_-_51553896 | 0.70 |
ENSMUST00000132734.1
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr11_+_53457184 | 0.70 |
ENSMUST00000109013.2
|
Shroom1
|
shroom family member 1 |
| chr2_+_153161303 | 0.69 |
ENSMUST00000089027.2
|
Tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr18_-_56562187 | 0.69 |
ENSMUST00000171844.2
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr13_+_64161862 | 0.69 |
ENSMUST00000021929.8
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr6_-_114921778 | 0.69 |
ENSMUST00000032459.7
|
Vgll4
|
vestigial like 4 (Drosophila) |
| chr11_+_53457273 | 0.68 |
ENSMUST00000093114.4
|
Shroom1
|
shroom family member 1 |
| chr13_-_54688065 | 0.68 |
ENSMUST00000125871.1
|
Rnf44
|
ring finger protein 44 |
| chr13_-_34077992 | 0.67 |
ENSMUST00000056427.8
|
Tubb2a
|
tubulin, beta 2A class IIA |
| chr1_+_95313607 | 0.66 |
ENSMUST00000059975.6
|
Fam174a
|
family with sequence similarity 174, member A |
| chr2_+_105668888 | 0.65 |
ENSMUST00000111086.4
ENSMUST00000111087.3 |
Pax6
|
paired box gene 6 |
| chr15_-_33687840 | 0.65 |
ENSMUST00000042021.3
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr18_-_80151467 | 0.65 |
ENSMUST00000066743.9
|
Adnp2
|
ADNP homeobox 2 |
| chr11_+_53457209 | 0.65 |
ENSMUST00000018531.5
|
Shroom1
|
shroom family member 1 |
| chr9_-_45955170 | 0.64 |
ENSMUST00000162072.1
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr13_-_54688246 | 0.64 |
ENSMUST00000122935.1
ENSMUST00000128257.1 |
Rnf44
|
ring finger protein 44 |
| chr11_+_94044331 | 0.64 |
ENSMUST00000024979.8
|
Spag9
|
sperm associated antigen 9 |
| chr18_-_56562215 | 0.63 |
ENSMUST00000170309.1
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr11_-_90390895 | 0.63 |
ENSMUST00000004051.7
|
Hlf
|
hepatic leukemia factor |
| chr11_+_94044241 | 0.63 |
ENSMUST00000103168.3
|
Spag9
|
sperm associated antigen 9 |
| chr17_+_23679363 | 0.63 |
ENSMUST00000024699.2
|
Cldn6
|
claudin 6 |
| chr16_+_13903152 | 0.62 |
ENSMUST00000128757.1
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr19_-_42128982 | 0.62 |
ENSMUST00000161873.1
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr2_+_156065180 | 0.62 |
ENSMUST00000038860.5
|
Spag4
|
sperm associated antigen 4 |
| chr18_-_56562261 | 0.62 |
ENSMUST00000066208.6
ENSMUST00000172734.1 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr11_+_79339792 | 0.61 |
ENSMUST00000108251.2
ENSMUST00000071325.2 |
Nf1
|
neurofibromatosis 1 |
| chr7_-_137314394 | 0.61 |
ENSMUST00000168203.1
ENSMUST00000106118.2 ENSMUST00000169486.2 ENSMUST00000033378.5 |
Ebf3
|
early B cell factor 3 |
| chr4_+_155847393 | 0.61 |
ENSMUST00000030948.9
ENSMUST00000168552.1 |
Dvl1
|
dishevelled, dsh homolog 1 (Drosophila) |
| chr1_-_92473801 | 0.60 |
ENSMUST00000027478.6
|
Ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 |
| chr13_-_53473074 | 0.59 |
ENSMUST00000021922.8
|
Msx2
|
msh homeobox 2 |
| chr11_-_43836243 | 0.59 |
ENSMUST00000167574.1
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr11_+_83302817 | 0.59 |
ENSMUST00000142680.1
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr4_+_140961203 | 0.58 |
ENSMUST00000010007.8
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr7_-_126949499 | 0.57 |
ENSMUST00000106339.1
ENSMUST00000052937.5 |
Asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr7_-_126898249 | 0.56 |
ENSMUST00000121532.1
ENSMUST00000032926.5 |
Tmem219
|
transmembrane protein 219 |
| chr7_+_132931142 | 0.56 |
ENSMUST00000106157.1
|
Zranb1
|
zinc finger, RAN-binding domain containing 1 |
| chr17_-_26508463 | 0.56 |
ENSMUST00000025025.6
|
Dusp1
|
dual specificity phosphatase 1 |
| chr2_-_173276144 | 0.56 |
ENSMUST00000139306.1
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_-_173276526 | 0.56 |
ENSMUST00000036248.6
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr11_-_119086221 | 0.55 |
ENSMUST00000026665.7
|
Cbx4
|
chromobox 4 |
| chr13_+_54621801 | 0.55 |
ENSMUST00000026991.9
ENSMUST00000137413.1 ENSMUST00000135232.1 ENSMUST00000124752.1 |
Faf2
|
Fas associated factor family member 2 |
| chr2_+_105668935 | 0.54 |
ENSMUST00000142772.1
|
Pax6
|
paired box gene 6 |
| chr19_-_42129043 | 0.54 |
ENSMUST00000018965.3
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr7_-_118584669 | 0.53 |
ENSMUST00000044195.4
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr11_-_100939457 | 0.53 |
ENSMUST00000138438.1
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr8_+_123477859 | 0.53 |
ENSMUST00000001520.7
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (yeast) |
| chr9_-_57467985 | 0.53 |
ENSMUST00000046587.6
|
Scamp5
|
secretory carrier membrane protein 5 |
| chr3_+_90248172 | 0.53 |
ENSMUST00000015467.8
|
Slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr15_+_102966794 | 0.52 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr6_-_134792596 | 0.51 |
ENSMUST00000100857.3
|
Dusp16
|
dual specificity phosphatase 16 |
| chr18_+_36348653 | 0.51 |
ENSMUST00000050584.3
|
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr16_-_4077778 | 0.51 |
ENSMUST00000006137.8
|
Trap1
|
TNF receptor-associated protein 1 |
| chr1_-_59237093 | 0.51 |
ENSMUST00000163058.1
ENSMUST00000027178.6 |
Als2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr11_+_94044194 | 0.50 |
ENSMUST00000092777.4
ENSMUST00000075695.6 |
Spag9
|
sperm associated antigen 9 |
| chr15_+_59648644 | 0.50 |
ENSMUST00000118228.1
|
Trib1
|
tribbles homolog 1 (Drosophila) |
| chr9_-_45955226 | 0.50 |
ENSMUST00000038488.9
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr13_-_54687696 | 0.50 |
ENSMUST00000177950.1
ENSMUST00000146931.1 |
Rnf44
|
ring finger protein 44 |
| chr9_+_64179289 | 0.49 |
ENSMUST00000034965.6
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr6_+_112273758 | 0.49 |
ENSMUST00000032376.5
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr10_+_96616998 | 0.49 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr11_+_110968056 | 0.49 |
ENSMUST00000125692.1
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr16_+_24393350 | 0.48 |
ENSMUST00000038053.6
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_+_52097737 | 0.48 |
ENSMUST00000040090.9
|
Nubpl
|
nucleotide binding protein-like |
| chr17_-_33760451 | 0.48 |
ENSMUST00000057373.7
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr4_-_95052188 | 0.47 |
ENSMUST00000107094.1
|
Jun
|
Jun oncogene |
| chr17_-_31855782 | 0.47 |
ENSMUST00000024839.4
|
Sik1
|
salt inducible kinase 1 |
| chr1_+_184034381 | 0.47 |
ENSMUST00000048655.7
|
Dusp10
|
dual specificity phosphatase 10 |
| chr7_-_30729505 | 0.47 |
ENSMUST00000006478.8
|
Tmem147
|
transmembrane protein 147 |
| chr2_-_160367057 | 0.47 |
ENSMUST00000099126.3
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| chr9_+_109054839 | 0.46 |
ENSMUST00000154184.1
|
Shisa5
|
shisa homolog 5 (Xenopus laevis) |
| chrX_-_48208566 | 0.46 |
ENSMUST00000037960.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr11_-_72796028 | 0.46 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr11_-_100939540 | 0.46 |
ENSMUST00000127638.1
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr12_+_75308308 | 0.46 |
ENSMUST00000118602.1
ENSMUST00000118966.1 ENSMUST00000055390.5 |
Rhoj
|
ras homolog gene family, member J |
| chr11_-_72795801 | 0.45 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr12_+_73286779 | 0.45 |
ENSMUST00000140523.1
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr18_+_38418946 | 0.45 |
ENSMUST00000025293.3
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr14_+_75242287 | 0.44 |
ENSMUST00000022576.8
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr4_-_109156610 | 0.44 |
ENSMUST00000161363.1
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr11_+_94044111 | 0.44 |
ENSMUST00000132079.1
|
Spag9
|
sperm associated antigen 9 |
| chr6_-_50566535 | 0.44 |
ENSMUST00000161401.1
|
Cycs
|
cytochrome c, somatic |
| chr7_-_30856178 | 0.44 |
ENSMUST00000094583.1
|
Ffar3
|
free fatty acid receptor 3 |
| chr3_+_82358056 | 0.44 |
ENSMUST00000091014.4
|
Map9
|
microtubule-associated protein 9 |
| chr15_+_81936911 | 0.43 |
ENSMUST00000135663.1
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr2_-_30205794 | 0.43 |
ENSMUST00000113663.2
ENSMUST00000044038.3 |
Ccbl1
|
cysteine conjugate-beta lyase 1 |
| chrX_+_143664365 | 0.43 |
ENSMUST00000126592.1
ENSMUST00000156449.1 ENSMUST00000155215.1 ENSMUST00000112865.1 |
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr3_-_146108047 | 0.43 |
ENSMUST00000160285.1
|
Wdr63
|
WD repeat domain 63 |
| chr15_-_73707387 | 0.43 |
ENSMUST00000064166.4
|
Gpr20
|
G protein-coupled receptor 20 |
| chr7_+_139389072 | 0.43 |
ENSMUST00000106098.1
ENSMUST00000026550.7 |
Inpp5a
|
inositol polyphosphate-5-phosphatase A |
| chr8_+_122775497 | 0.43 |
ENSMUST00000015160.5
|
Acsf3
|
acyl-CoA synthetase family member 3 |
| chr17_+_36958571 | 0.42 |
ENSMUST00000040177.6
|
Znrd1as
|
Znrd1 antisense |
| chr4_+_43983496 | 0.42 |
ENSMUST00000095107.1
|
Ccin
|
calicin |
| chr11_+_101984270 | 0.42 |
ENSMUST00000176722.1
ENSMUST00000175972.1 |
1700006E09Rik
|
RIKEN cDNA 1700006E09 gene |
| chr9_+_109054903 | 0.42 |
ENSMUST00000151141.1
ENSMUST00000152771.1 |
Shisa5
|
shisa homolog 5 (Xenopus laevis) |
| chr5_-_25223153 | 0.42 |
ENSMUST00000066954.1
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene |
| chr17_+_28177339 | 0.42 |
ENSMUST00000073534.2
ENSMUST00000002318.1 |
Zfp523
|
zinc finger protein 523 |
| chr8_+_4325205 | 0.41 |
ENSMUST00000069762.9
ENSMUST00000098949.4 ENSMUST00000127460.1 ENSMUST00000136191.1 |
Ccl25
Ccl25
|
chemokine (C-C motif) ligand 25 chemokine (C-C motif) ligand 25 |
| chr13_+_113035111 | 0.41 |
ENSMUST00000180543.1
ENSMUST00000181568.1 ENSMUST00000109244.2 ENSMUST00000181117.1 ENSMUST00000181741.1 |
Cdc20b
|
cell division cycle 20B |
| chr4_-_95052170 | 0.41 |
ENSMUST00000058555.2
|
Jun
|
Jun oncogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Crem_Jdp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.7 | 2.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.7 | 2.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.6 | 1.9 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.6 | 1.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) cellular response to mercury ion(GO:0071288) |
| 0.6 | 6.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 2.5 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 | 1.4 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.4 | 1.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 1.9 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.4 | 1.4 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.3 | 1.4 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.3 | 0.9 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.3 | 1.5 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.3 | 0.9 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 2.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 0.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 1.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.8 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 1.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.2 | 1.2 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.2 | 1.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 2.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 0.4 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 1.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 1.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 2.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 1.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 0.6 | GO:0051795 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) |
| 0.2 | 0.6 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.2 | 1.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.6 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.7 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 0.5 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.5 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 0.5 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 | 2.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 2.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 0.9 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 | 0.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.4 | GO:0002879 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 8.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 1.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.6 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.8 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.3 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 0.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 2.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 1.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 1.5 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.3 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.4 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.3 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.7 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.5 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0090467 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.3 | GO:0048597 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 4.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.5 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 1.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.8 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 1.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.8 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.2 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0071338 | positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.3 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.3 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.4 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.4 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 0.4 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.5 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0060067 | cervix development(GO:0060067) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.6 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.7 | 2.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.6 | 1.8 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 1.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 4.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 2.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 9.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 5.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 2.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 1.9 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 1.4 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.4 | 2.4 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 2.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 1.8 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.3 | 1.5 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 2.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.2 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.2 | 2.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 1.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 2.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 3.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 1.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 3.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 8.6 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |