Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
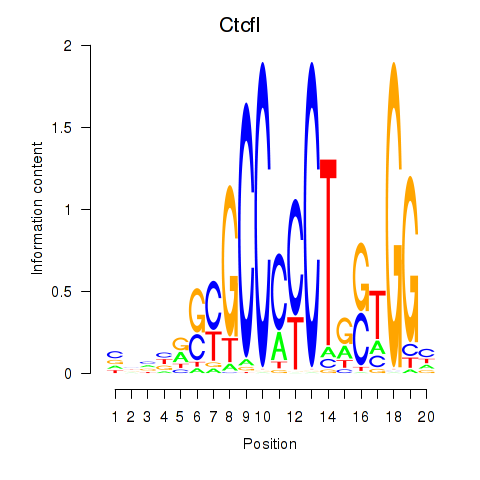
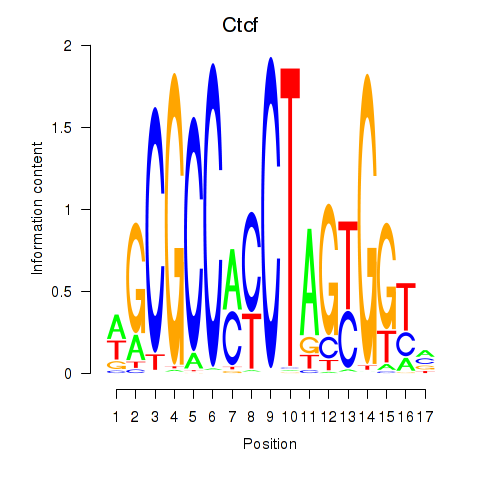
Results for Ctcfl_Ctcf
Z-value: 0.76
Transcription factors associated with Ctcfl_Ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ctcfl
|
ENSMUSG00000070495.5 | CCCTC-binding factor (zinc finger protein)-like |
|
Ctcf
|
ENSMUSG00000005698.9 | CCCTC-binding factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcfl | mm10_v2_chr2_-_173119402_173119525 | -0.45 | 5.9e-03 | Click! |
| Ctcf | mm10_v2_chr8_+_105636509_105636589 | 0.08 | 6.6e-01 | Click! |
Activity profile of Ctcfl_Ctcf motif
Sorted Z-values of Ctcfl_Ctcf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_32286946 | 4.21 |
ENSMUST00000101387.3
|
Hbq1b
|
hemoglobin, theta 1B |
| chr19_-_45816007 | 2.76 |
ENSMUST00000079431.3
ENSMUST00000026247.6 ENSMUST00000162528.2 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr11_+_32300069 | 2.48 |
ENSMUST00000020535.1
|
Hbq1a
|
hemoglobin, theta 1A |
| chr7_-_109170308 | 2.20 |
ENSMUST00000036992.7
|
Lmo1
|
LIM domain only 1 |
| chr2_+_122637867 | 2.14 |
ENSMUST00000110512.3
|
AA467197
|
expressed sequence AA467197 |
| chr17_+_26917091 | 1.94 |
ENSMUST00000078961.4
|
Kifc5b
|
kinesin family member C5B |
| chr5_-_137741601 | 1.90 |
ENSMUST00000119498.1
ENSMUST00000061789.7 |
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr2_+_122637844 | 1.90 |
ENSMUST00000047498.8
|
AA467197
|
expressed sequence AA467197 |
| chr5_-_5749317 | 1.83 |
ENSMUST00000015796.2
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr5_+_144768536 | 1.81 |
ENSMUST00000128550.1
|
Trrap
|
transformation/transcription domain-associated protein |
| chr14_+_33319703 | 1.80 |
ENSMUST00000111955.1
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr5_-_36398090 | 1.70 |
ENSMUST00000037370.7
ENSMUST00000070720.6 |
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr5_+_149678224 | 1.59 |
ENSMUST00000100404.3
|
B3galtl
|
beta 1,3-galactosyltransferase-like |
| chr16_-_20426375 | 1.58 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_+_154613297 | 1.58 |
ENSMUST00000081926.6
ENSMUST00000109702.1 |
Zfp341
|
zinc finger protein 341 |
| chr5_-_137741102 | 1.58 |
ENSMUST00000149512.1
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr9_-_44234014 | 1.41 |
ENSMUST00000037644.6
|
Cbl
|
Casitas B-lineage lymphoma |
| chr11_-_76577701 | 1.38 |
ENSMUST00000176179.1
|
Abr
|
active BCR-related gene |
| chr13_-_53286052 | 1.34 |
ENSMUST00000021918.8
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr17_-_25433775 | 1.33 |
ENSMUST00000159610.1
ENSMUST00000159048.1 ENSMUST00000078496.5 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr16_-_20426322 | 1.32 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_+_91366412 | 1.32 |
ENSMUST00000086861.5
|
Fam132b
|
family with sequence similarity 132, member B |
| chr7_+_122159422 | 1.32 |
ENSMUST00000033154.6
|
Plk1
|
polo-like kinase 1 |
| chr16_-_20425881 | 1.32 |
ENSMUST00000077867.3
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_+_45434833 | 1.31 |
ENSMUST00000003964.8
|
Gys1
|
glycogen synthase 1, muscle |
| chr2_-_150668198 | 1.31 |
ENSMUST00000028944.3
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr2_+_29619692 | 1.30 |
ENSMUST00000095087.4
ENSMUST00000091146.5 ENSMUST00000102872.4 |
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr3_-_96708524 | 1.27 |
ENSMUST00000029742.5
ENSMUST00000171249.1 |
Nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr2_-_32353247 | 1.26 |
ENSMUST00000078352.5
ENSMUST00000113352.2 ENSMUST00000113365.1 |
Dnm1
|
dynamin 1 |
| chr9_+_65101453 | 1.24 |
ENSMUST00000077696.6
ENSMUST00000035499.4 ENSMUST00000166273.1 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr7_+_45434876 | 1.24 |
ENSMUST00000107766.1
|
Gys1
|
glycogen synthase 1, muscle |
| chr15_-_102350692 | 1.23 |
ENSMUST00000041208.7
|
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr4_+_140701466 | 1.23 |
ENSMUST00000038893.5
ENSMUST00000138808.1 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr10_+_3973086 | 1.22 |
ENSMUST00000117291.1
ENSMUST00000120585.1 ENSMUST00000043735.7 |
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr2_-_32353283 | 1.22 |
ENSMUST00000091089.5
ENSMUST00000113350.1 |
Dnm1
|
dynamin 1 |
| chr7_-_67803489 | 1.21 |
ENSMUST00000181235.1
|
4833412C05Rik
|
RIKEN cDNA 4833412C05 gene |
| chr17_-_56036546 | 1.19 |
ENSMUST00000003268.9
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr14_+_53806497 | 1.19 |
ENSMUST00000103672.4
|
Trav17
|
T cell receptor alpha variable 17 |
| chr9_-_108452377 | 1.18 |
ENSMUST00000035232.7
|
Klhdc8b
|
kelch domain containing 8B |
| chr19_+_7417586 | 1.17 |
ENSMUST00000159348.1
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr17_-_33890539 | 1.16 |
ENSMUST00000173386.1
|
Kifc1
|
kinesin family member C1 |
| chr17_-_33890584 | 1.14 |
ENSMUST00000114361.2
ENSMUST00000173492.1 |
Kifc1
|
kinesin family member C1 |
| chr5_-_123132651 | 1.12 |
ENSMUST00000031401.5
|
Rhof
|
ras homolog gene family, member f |
| chr5_+_23850590 | 1.11 |
ENSMUST00000181574.1
|
2700038G22Rik
|
RIKEN cDNA 2700038G22 gene |
| chr11_-_53470479 | 1.08 |
ENSMUST00000057722.2
|
Gm9837
|
predicted gene 9837 |
| chr7_-_13009795 | 1.06 |
ENSMUST00000051390.7
ENSMUST00000172240.1 |
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr7_-_15967470 | 1.06 |
ENSMUST00000144956.1
ENSMUST00000098799.3 |
Ehd2
|
EH-domain containing 2 |
| chr8_-_122678653 | 1.06 |
ENSMUST00000134045.1
|
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr1_-_132367879 | 1.05 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr16_+_31878810 | 1.02 |
ENSMUST00000023464.5
|
Mfi2
|
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr11_+_61956779 | 1.00 |
ENSMUST00000049836.7
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_-_73184840 | 0.99 |
ENSMUST00000044113.10
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr4_-_154025657 | 0.96 |
ENSMUST00000146426.1
|
Smim1
|
small integral membrane protein 1 |
| chr1_-_191575534 | 0.95 |
ENSMUST00000027933.5
|
Dtl
|
denticleless homolog (Drosophila) |
| chr8_-_122551316 | 0.95 |
ENSMUST00000067252.7
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr4_-_154026037 | 0.93 |
ENSMUST00000131325.2
ENSMUST00000146054.1 ENSMUST00000126119.1 ENSMUST00000125533.2 |
Smim1
|
small integral membrane protein 1 |
| chr4_-_141078302 | 0.93 |
ENSMUST00000030760.8
|
Necap2
|
NECAP endocytosis associated 2 |
| chr2_-_91963507 | 0.92 |
ENSMUST00000028667.3
|
Dgkz
|
diacylglycerol kinase zeta |
| chr6_+_42264983 | 0.91 |
ENSMUST00000031895.6
|
Casp2
|
caspase 2 |
| chr10_-_80561528 | 0.90 |
ENSMUST00000057910.9
|
Rexo1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr4_-_149137536 | 0.89 |
ENSMUST00000176124.1
ENSMUST00000177408.1 ENSMUST00000105695.1 ENSMUST00000030813.3 |
Apitd1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr18_-_80363622 | 0.89 |
ENSMUST00000184366.1
|
Kcng2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr4_-_154025867 | 0.88 |
ENSMUST00000130175.1
ENSMUST00000182151.1 |
Smim1
|
small integral membrane protein 1 |
| chr7_-_28372233 | 0.83 |
ENSMUST00000094644.4
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_+_3514861 | 0.82 |
ENSMUST00000094469.4
|
Selm
|
selenoprotein M |
| chr7_-_28372597 | 0.82 |
ENSMUST00000144700.1
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_-_107425029 | 0.82 |
ENSMUST00000003946.8
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr7_-_45434590 | 0.80 |
ENSMUST00000107771.3
ENSMUST00000141761.1 |
Ruvbl2
|
RuvB-like protein 2 |
| chr4_-_154025926 | 0.79 |
ENSMUST00000132541.1
ENSMUST00000143047.1 |
Smim1
|
small integral membrane protein 1 |
| chr9_+_21546842 | 0.79 |
ENSMUST00000034703.8
ENSMUST00000115395.3 ENSMUST00000115394.1 |
Carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr7_-_28372494 | 0.78 |
ENSMUST00000119990.1
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chrX_-_133898399 | 0.78 |
ENSMUST00000087557.5
|
Tspan6
|
tetraspanin 6 |
| chr9_+_110857958 | 0.77 |
ENSMUST00000051097.1
|
Prss50
|
protease, serine, 50 |
| chr8_+_70282978 | 0.77 |
ENSMUST00000110124.2
|
Homer3
|
homer homolog 3 (Drosophila) |
| chr7_+_108934405 | 0.77 |
ENSMUST00000033342.6
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr11_+_101733011 | 0.77 |
ENSMUST00000129741.1
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr15_-_76817970 | 0.75 |
ENSMUST00000175843.1
ENSMUST00000177026.1 ENSMUST00000176736.1 ENSMUST00000036176.8 ENSMUST00000176219.1 ENSMUST00000077821.3 |
Arhgap39
|
Rho GTPase activating protein 39 |
| chr9_+_110333402 | 0.75 |
ENSMUST00000133114.1
ENSMUST00000125759.1 |
Scap
|
SREBF chaperone |
| chr7_-_45136231 | 0.74 |
ENSMUST00000124300.1
ENSMUST00000085377.5 |
Rpl13a
Flt3l
|
ribosomal protein L13A FMS-like tyrosine kinase 3 ligand |
| chr9_+_66158206 | 0.74 |
ENSMUST00000034944.2
|
Dapk2
|
death-associated protein kinase 2 |
| chr10_-_18546049 | 0.74 |
ENSMUST00000020000.5
|
Hebp2
|
heme binding protein 2 |
| chr19_+_46304709 | 0.73 |
ENSMUST00000073116.5
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr5_-_114823460 | 0.72 |
ENSMUST00000140374.1
ENSMUST00000100850.4 |
Gm20499
2610524H06Rik
|
predicted gene 20499 RIKEN cDNA 2610524H06 gene |
| chr18_+_73863672 | 0.72 |
ENSMUST00000134847.1
|
Mro
|
maestro |
| chr6_+_87887814 | 0.72 |
ENSMUST00000113607.3
ENSMUST00000049966.5 |
Copg1
|
coatomer protein complex, subunit gamma 1 |
| chrX_-_133898292 | 0.71 |
ENSMUST00000176718.1
ENSMUST00000176641.1 |
Tspan6
|
tetraspanin 6 |
| chr18_+_68337504 | 0.70 |
ENSMUST00000172148.1
|
Mc5r
|
melanocortin 5 receptor |
| chrX_-_134751331 | 0.70 |
ENSMUST00000113194.1
ENSMUST00000052431.5 |
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr3_-_95217741 | 0.70 |
ENSMUST00000107204.1
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr4_+_152039315 | 0.69 |
ENSMUST00000084116.6
ENSMUST00000105663.1 ENSMUST00000103197.3 |
Nol9
|
nucleolar protein 9 |
| chr12_-_101028983 | 0.69 |
ENSMUST00000068411.3
ENSMUST00000085096.3 |
Ccdc88c
|
coiled-coil domain containing 88C |
| chr17_-_7827289 | 0.69 |
ENSMUST00000167580.1
ENSMUST00000169126.1 |
Fndc1
|
fibronectin type III domain containing 1 |
| chr10_-_88379051 | 0.68 |
ENSMUST00000138159.1
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr4_-_43025756 | 0.68 |
ENSMUST00000098109.2
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr11_-_69579320 | 0.68 |
ENSMUST00000048139.5
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chrX_+_106027300 | 0.67 |
ENSMUST00000055941.6
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr1_+_172482199 | 0.67 |
ENSMUST00000135267.1
ENSMUST00000052629.6 ENSMUST00000111235.2 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr4_+_151933691 | 0.67 |
ENSMUST00000062904.4
|
Dnajc11
|
DnaJ (Hsp40) homolog, subfamily C, member 11 |
| chr2_+_79255500 | 0.67 |
ENSMUST00000099972.4
|
Itga4
|
integrin alpha 4 |
| chr16_-_10785525 | 0.66 |
ENSMUST00000038099.4
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr2_-_114175274 | 0.66 |
ENSMUST00000102543.4
|
Aqr
|
aquarius |
| chr10_-_80577285 | 0.66 |
ENSMUST00000038558.8
|
Klf16
|
Kruppel-like factor 16 |
| chr7_-_46710642 | 0.66 |
ENSMUST00000143082.1
|
Saal1
|
serum amyloid A-like 1 |
| chr11_+_63132569 | 0.66 |
ENSMUST00000108701.1
|
Pmp22
|
peripheral myelin protein 22 |
| chr11_+_63133068 | 0.66 |
ENSMUST00000108700.1
|
Pmp22
|
peripheral myelin protein 22 |
| chr11_+_84179792 | 0.65 |
ENSMUST00000137500.2
ENSMUST00000130012.2 |
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr10_+_128459236 | 0.65 |
ENSMUST00000105235.2
ENSMUST00000099131.3 ENSMUST00000026433.7 |
Smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr6_+_83165920 | 0.65 |
ENSMUST00000077407.5
ENSMUST00000113913.1 ENSMUST00000130212.1 |
Dctn1
|
dynactin 1 |
| chr11_-_75169519 | 0.65 |
ENSMUST00000055619.4
|
Hic1
|
hypermethylated in cancer 1 |
| chr19_-_10304867 | 0.64 |
ENSMUST00000039327.4
|
Dagla
|
diacylglycerol lipase, alpha |
| chr9_-_60522017 | 0.63 |
ENSMUST00000140824.1
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr11_-_101175440 | 0.63 |
ENSMUST00000062759.3
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr11_-_62648458 | 0.63 |
ENSMUST00000057194.8
|
Fam211a
|
family with sequence similarity 211, member A |
| chr1_+_82839449 | 0.63 |
ENSMUST00000113444.1
ENSMUST00000063380.4 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr11_-_59964936 | 0.63 |
ENSMUST00000062405.7
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chrX_+_106027259 | 0.63 |
ENSMUST00000113557.1
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr5_-_143315360 | 0.62 |
ENSMUST00000046418.2
|
E130309D02Rik
|
RIKEN cDNA E130309D02 gene |
| chr4_-_116167591 | 0.61 |
ENSMUST00000030465.3
ENSMUST00000143426.1 |
Tspan1
|
tetraspanin 1 |
| chr19_-_24280551 | 0.61 |
ENSMUST00000081333.4
|
Fxn
|
frataxin |
| chr9_-_102354685 | 0.61 |
ENSMUST00000035129.7
ENSMUST00000085169.5 |
Ephb1
|
Eph receptor B1 |
| chr14_+_57999305 | 0.61 |
ENSMUST00000180534.1
|
3110083C13Rik
|
RIKEN cDNA 3110083C13 gene |
| chr19_-_4283033 | 0.60 |
ENSMUST00000167215.1
ENSMUST00000056888.6 |
Ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr10_-_86022325 | 0.59 |
ENSMUST00000181665.1
|
A230060F14Rik
|
RIKEN cDNA A230060F14 gene |
| chr3_-_87768966 | 0.59 |
ENSMUST00000174776.1
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr11_+_43528759 | 0.59 |
ENSMUST00000050574.6
|
Ccnjl
|
cyclin J-like |
| chr13_-_113046357 | 0.59 |
ENSMUST00000022282.3
|
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr12_-_27342696 | 0.58 |
ENSMUST00000079063.5
|
Sox11
|
SRY-box containing gene 11 |
| chr4_+_48540067 | 0.58 |
ENSMUST00000064807.2
|
Msantd3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr6_-_89362581 | 0.57 |
ENSMUST00000163139.1
|
Plxna1
|
plexin A1 |
| chr11_+_101732950 | 0.57 |
ENSMUST00000039152.7
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr4_-_116464151 | 0.57 |
ENSMUST00000106486.1
ENSMUST00000106485.1 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_157914618 | 0.57 |
ENSMUST00000109523.1
|
Vstm2l
|
V-set and transmembrane domain containing 2-like |
| chr1_+_135132693 | 0.56 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr4_-_131838231 | 0.56 |
ENSMUST00000030741.2
ENSMUST00000105987.2 |
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chr9_-_96889381 | 0.56 |
ENSMUST00000112951.2
ENSMUST00000126411.1 ENSMUST00000078478.1 ENSMUST00000119141.1 ENSMUST00000120101.1 |
Acpl2
|
acid phosphatase-like 2 |
| chr12_+_99964499 | 0.55 |
ENSMUST00000177549.1
ENSMUST00000160413.1 ENSMUST00000162221.1 ENSMUST00000049788.8 |
Kcnk13
|
potassium channel, subfamily K, member 13 |
| chr19_+_5568002 | 0.54 |
ENSMUST00000096318.3
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr12_+_24708984 | 0.53 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_-_127030813 | 0.53 |
ENSMUST00000040560.4
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr2_-_114175321 | 0.53 |
ENSMUST00000043160.6
|
Aqr
|
aquarius |
| chr4_-_43045686 | 0.53 |
ENSMUST00000107956.1
ENSMUST00000107957.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr19_+_6400523 | 0.53 |
ENSMUST00000146831.1
ENSMUST00000035716.8 ENSMUST00000138555.1 ENSMUST00000167240.1 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr2_-_164911586 | 0.53 |
ENSMUST00000041361.7
|
Zfp335
|
zinc finger protein 335 |
| chr19_-_6141402 | 0.52 |
ENSMUST00000025893.5
|
Arl2
|
ADP-ribosylation factor-like 2 |
| chr2_-_152831665 | 0.52 |
ENSMUST00000156688.1
ENSMUST00000007803.5 |
Bcl2l1
|
BCL2-like 1 |
| chr19_-_6015769 | 0.52 |
ENSMUST00000164843.1
|
Capn1
|
calpain 1 |
| chr17_-_24479034 | 0.52 |
ENSMUST00000179163.1
ENSMUST00000070888.6 |
Mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr9_+_110333276 | 0.51 |
ENSMUST00000125823.1
ENSMUST00000131328.1 |
Scap
|
SREBF chaperone |
| chr7_+_25686994 | 0.51 |
ENSMUST00000002678.9
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr5_+_115559505 | 0.51 |
ENSMUST00000156359.1
ENSMUST00000152976.1 |
Rplp0
|
ribosomal protein, large, P0 |
| chr11_-_102447647 | 0.51 |
ENSMUST00000049057.4
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr9_-_106887000 | 0.51 |
ENSMUST00000055843.7
|
Rbm15b
|
RNA binding motif protein 15B |
| chr12_+_80518990 | 0.51 |
ENSMUST00000021558.6
|
Galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr1_+_34459746 | 0.50 |
ENSMUST00000027302.7
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr7_+_66839752 | 0.50 |
ENSMUST00000107478.1
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr4_-_43046196 | 0.50 |
ENSMUST00000036462.5
|
Fam214b
|
family with sequence similarity 214, member B |
| chr15_-_99705490 | 0.50 |
ENSMUST00000163472.2
|
Gm17349
|
predicted gene, 17349 |
| chr5_+_137641334 | 0.50 |
ENSMUST00000177466.1
ENSMUST00000166099.2 |
Sap25
|
sin3 associated polypeptide |
| chr11_+_62648811 | 0.50 |
ENSMUST00000127589.1
ENSMUST00000155759.2 |
Mmgt2
|
membrane magnesium transporter 2 |
| chr7_+_31059342 | 0.50 |
ENSMUST00000039775.7
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr8_-_105565985 | 0.50 |
ENSMUST00000013304.7
|
Atp6v0d1
|
ATPase, H+ transporting, lysosomal V0 subunit D1 |
| chr5_+_115559467 | 0.49 |
ENSMUST00000086519.5
|
Rplp0
|
ribosomal protein, large, P0 |
| chr7_+_16098458 | 0.49 |
ENSMUST00000006181.6
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chr1_+_172481788 | 0.49 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr9_-_42944479 | 0.48 |
ENSMUST00000114865.1
|
Grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr10_-_127030789 | 0.48 |
ENSMUST00000120547.1
ENSMUST00000152054.1 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr10_+_121033960 | 0.47 |
ENSMUST00000020439.4
ENSMUST00000175867.1 |
Wif1
|
Wnt inhibitory factor 1 |
| chr1_+_74125535 | 0.47 |
ENSMUST00000080167.4
ENSMUST00000127134.1 |
Rufy4
|
RUN and FYVE domain containing 4 |
| chr2_+_3118335 | 0.47 |
ENSMUST00000115099.2
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr19_-_4306214 | 0.47 |
ENSMUST00000171123.1
ENSMUST00000088737.4 |
Adrbk1
|
adrenergic receptor kinase, beta 1 |
| chr2_+_25403044 | 0.46 |
ENSMUST00000071442.5
|
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr16_+_17276662 | 0.46 |
ENSMUST00000069420.4
|
Tmem191c
|
transmembrane protein 191C |
| chr6_-_41636389 | 0.46 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr15_-_44788016 | 0.46 |
ENSMUST00000090057.4
ENSMUST00000110269.1 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr2_+_65620829 | 0.46 |
ENSMUST00000028377.7
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1 |
| chr2_-_119271202 | 0.46 |
ENSMUST00000037360.7
|
Rhov
|
ras homolog gene family, member V |
| chr5_-_139736291 | 0.46 |
ENSMUST00000044642.10
|
Micall2
|
MICAL-like 2 |
| chr12_+_24708241 | 0.45 |
ENSMUST00000020980.5
|
Rrm2
|
ribonucleotide reductase M2 |
| chr4_-_43025792 | 0.44 |
ENSMUST00000067481.5
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chrX_-_53240101 | 0.44 |
ENSMUST00000074861.2
|
Plac1
|
placental specific protein 1 |
| chr11_+_62648656 | 0.44 |
ENSMUST00000062860.4
|
Mmgt2
|
membrane magnesium transporter 2 |
| chr9_+_71215779 | 0.44 |
ENSMUST00000034723.5
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr4_-_132345715 | 0.44 |
ENSMUST00000084250.4
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr14_+_53616315 | 0.44 |
ENSMUST00000180972.1
ENSMUST00000185023.1 |
Trav12-2
|
T cell receptor alpha variable 12-2 |
| chr5_+_136116631 | 0.43 |
ENSMUST00000111127.1
ENSMUST00000041366.7 ENSMUST00000111129.1 |
Polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr7_+_66839726 | 0.43 |
ENSMUST00000098382.3
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr3_+_95622236 | 0.43 |
ENSMUST00000074353.4
|
Rps10-ps1
|
ribosomal protein S10, pseudogene 1 |
| chr2_-_152831112 | 0.43 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr19_+_46056539 | 0.42 |
ENSMUST00000111899.1
ENSMUST00000099392.3 ENSMUST00000062322.4 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr8_+_94857450 | 0.42 |
ENSMUST00000109521.3
|
Polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr1_-_136234113 | 0.42 |
ENSMUST00000120339.1
ENSMUST00000048668.8 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr11_-_45944910 | 0.42 |
ENSMUST00000129820.1
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr6_+_127446819 | 0.42 |
ENSMUST00000112191.1
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr8_+_88272403 | 0.42 |
ENSMUST00000169037.1
|
Adcy7
|
adenylate cyclase 7 |
| chr8_-_105484350 | 0.42 |
ENSMUST00000044286.5
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr1_+_153749496 | 0.42 |
ENSMUST00000182722.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_+_135198034 | 0.42 |
ENSMUST00000130612.1
|
8430419L09Rik
|
RIKEN cDNA 8430419L09 gene |
| chr19_-_4305955 | 0.41 |
ENSMUST00000025791.5
|
Adrbk1
|
adrenergic receptor kinase, beta 1 |
| chr10_+_86021961 | 0.41 |
ENSMUST00000130320.1
|
Fbxo7
|
F-box protein 7 |
| chr1_-_38129618 | 0.41 |
ENSMUST00000027251.6
|
Rev1
|
REV1 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ctcfl_Ctcf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 0.6 | 2.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.5 | 2.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 1.3 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.4 | 2.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 1.3 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 0.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 1.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.3 | 0.8 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.3 | 1.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.3 | 1.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 1.0 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.7 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 2.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.9 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 0.7 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 1.3 | GO:0051790 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.8 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.2 | 1.0 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 0.2 | 1.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 0.6 | GO:0019230 | proprioception(GO:0019230) |
| 0.2 | 1.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 0.8 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 0.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.2 | 0.7 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 1.7 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.2 | 0.7 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 0.7 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.2 | 0.6 | GO:0098917 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.2 | 0.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.7 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.7 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.9 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.1 | 1.3 | GO:0034651 | cortisol biosynthetic process(GO:0034651) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.7 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.1 | 1.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 1.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 1.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.8 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) positive regulation of mitophagy(GO:1903599) |
| 0.1 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0048320 | axial mesoderm formation(GO:0048320) chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.7 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 0.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.5 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 | 1.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.4 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.7 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.2 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.1 | 0.4 | GO:0042905 | ureter maturation(GO:0035799) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.2 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.1 | 0.2 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.1 | GO:0072319 | vesicle uncoating(GO:0072319) |
| 0.1 | 1.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.3 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.1 | 0.2 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 1.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.4 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 1.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.5 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.0 | 0.3 | GO:2001268 | hematopoietic stem cell homeostasis(GO:0061484) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.6 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.4 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 2.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 1.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.7 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 3.8 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.6 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 1.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.5 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 0.7 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 2.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 4.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 0.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 1.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 2.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.3 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 1.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 2.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.2 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.2 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 0.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.9 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 3.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.6 | 2.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 1.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 1.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 1.3 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 0.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 1.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.3 | 1.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 1.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 2.4 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 4.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.9 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.1 | 0.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.4 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.9 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.7 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.6 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 2.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.2 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.9 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.4 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 0.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.2 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 1.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 4.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 3.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 1.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 3.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |