Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Ehf
Z-value: 0.26
Transcription factors associated with Ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ehf
|
ENSMUSG00000012350.9 | ets homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ehf | mm10_v2_chr2_-_103303158_103303171 | -0.26 | 1.2e-01 | Click! |
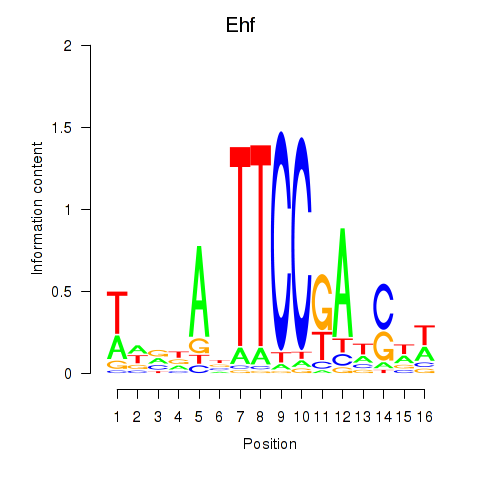
Activity profile of Ehf motif
Sorted Z-values of Ehf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_123289862 | 0.75 |
ENSMUST00000032239.4
ENSMUST00000177367.1 |
Clec4e
|
C-type lectin domain family 4, member e |
| chr3_+_28781305 | 0.69 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr17_+_34377124 | 0.41 |
ENSMUST00000080254.5
|
Btnl1
|
butyrophilin-like 1 |
| chr2_+_164805082 | 0.38 |
ENSMUST00000052107.4
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr17_-_40794063 | 0.37 |
ENSMUST00000131699.1
ENSMUST00000024724.7 ENSMUST00000144243.1 |
Crisp2
|
cysteine-rich secretory protein 2 |
| chr15_-_73184840 | 0.35 |
ENSMUST00000044113.10
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr18_-_24603791 | 0.26 |
ENSMUST00000070726.3
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr18_-_24603464 | 0.25 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr7_-_45239108 | 0.24 |
ENSMUST00000033063.6
|
Cd37
|
CD37 antigen |
| chr11_+_3514861 | 0.22 |
ENSMUST00000094469.4
|
Selm
|
selenoprotein M |
| chr3_+_116594959 | 0.22 |
ENSMUST00000029571.8
|
Sass6
|
spindle assembly 6 homolog (C. elegans) |
| chr3_-_95855753 | 0.21 |
ENSMUST00000161476.1
|
Prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr3_-_95855860 | 0.21 |
ENSMUST00000015892.7
|
Prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr8_-_104641666 | 0.19 |
ENSMUST00000093234.4
|
Fam96b
|
family with sequence similarity 96, member B |
| chr1_-_153549697 | 0.18 |
ENSMUST00000041874.7
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chrX_+_71663665 | 0.17 |
ENSMUST00000070449.5
|
Gpr50
|
G-protein-coupled receptor 50 |
| chr4_-_64276595 | 0.17 |
ENSMUST00000141162.1
|
Gm11217
|
predicted gene 11217 |
| chr6_+_142413441 | 0.15 |
ENSMUST00000088263.4
|
B230216G23Rik
|
RIKEN cDNA B230216G23 gene |
| chr8_-_105471481 | 0.15 |
ENSMUST00000014990.6
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr13_+_109260481 | 0.14 |
ENSMUST00000153234.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_+_142414012 | 0.13 |
ENSMUST00000141548.1
|
B230216G23Rik
|
RIKEN cDNA B230216G23 gene |
| chr2_-_94438081 | 0.12 |
ENSMUST00000028617.6
|
Api5
|
apoptosis inhibitor 5 |
| chr2_+_104122738 | 0.09 |
ENSMUST00000040374.5
|
A930018P22Rik
|
RIKEN cDNA A930018P22 gene |
| chr8_+_54600774 | 0.09 |
ENSMUST00000033917.6
|
Spata4
|
spermatogenesis associated 4 |
| chr6_-_30915552 | 0.09 |
ENSMUST00000048580.1
|
Tsga13
|
testis specific gene A13 |
| chr9_-_106447584 | 0.08 |
ENSMUST00000171678.1
ENSMUST00000048685.6 ENSMUST00000171925.1 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr11_+_72689997 | 0.07 |
ENSMUST00000155998.1
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr2_-_164804876 | 0.07 |
ENSMUST00000103094.4
ENSMUST00000017451.6 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr14_+_53640701 | 0.05 |
ENSMUST00000179267.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr16_-_19286837 | 0.05 |
ENSMUST00000056727.4
|
Olfr164
|
olfactory receptor 164 |
| chrX_+_153558591 | 0.05 |
ENSMUST00000112573.1
ENSMUST00000056754.3 |
Cypt3
|
cysteine-rich perinuclear theca 3 |
| chr2_+_129100995 | 0.05 |
ENSMUST00000103205.4
ENSMUST00000028874.7 |
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr13_-_8870999 | 0.04 |
ENSMUST00000177404.1
ENSMUST00000176922.1 ENSMUST00000021572.4 |
Wdr37
|
WD repeat domain 37 |
| chr19_+_6164433 | 0.02 |
ENSMUST00000045042.7
|
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr16_+_16302955 | 0.01 |
ENSMUST00000159962.1
ENSMUST00000059955.8 |
Yars2
|
tyrosyl-tRNA synthetase 2 (mitochondrial) |
| chr1_-_138856819 | 0.01 |
ENSMUST00000112025.1
|
2310009B15Rik
|
RIKEN cDNA 2310009B15 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ehf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.4 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |