Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
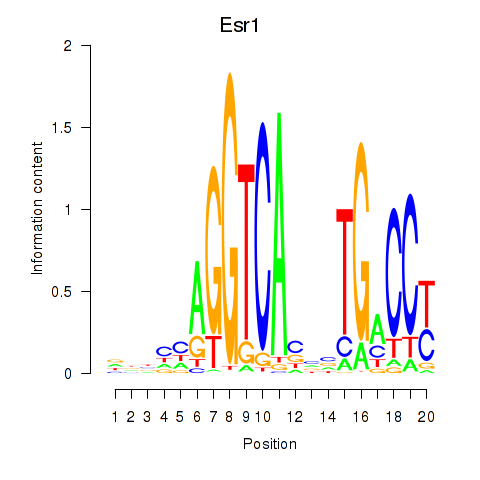
Results for Esr1
Z-value: 0.81
Transcription factors associated with Esr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esr1
|
ENSMUSG00000019768.10 | estrogen receptor 1 (alpha) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esr1 | mm10_v2_chr10_+_4611971_4612021 | -0.25 | 1.4e-01 | Click! |
Activity profile of Esr1 motif
Sorted Z-values of Esr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_58918004 | 2.09 |
ENSMUST00000108818.3
ENSMUST00000020792.5 |
Btnl10
|
butyrophilin-like 10 |
| chr6_-_69243445 | 2.05 |
ENSMUST00000101325.3
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr19_-_7711263 | 1.78 |
ENSMUST00000025666.7
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr11_+_87794206 | 1.76 |
ENSMUST00000121303.1
|
Mpo
|
myeloperoxidase |
| chr9_+_111019284 | 1.72 |
ENSMUST00000035077.3
|
Ltf
|
lactotransferrin |
| chr11_-_83286722 | 1.70 |
ENSMUST00000163961.2
|
Slfn14
|
schlafen family member 14 |
| chr11_+_87793470 | 1.66 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr1_+_131638485 | 1.66 |
ENSMUST00000112411.1
|
Ctse
|
cathepsin E |
| chr10_+_79886302 | 1.64 |
ENSMUST00000046091.5
|
Elane
|
elastase, neutrophil expressed |
| chr1_+_131638306 | 1.59 |
ENSMUST00000073350.6
|
Ctse
|
cathepsin E |
| chr14_-_56085214 | 1.49 |
ENSMUST00000015594.7
|
Mcpt8
|
mast cell protease 8 |
| chr17_+_25471564 | 1.44 |
ENSMUST00000025002.1
|
Tekt4
|
tektin 4 |
| chr4_-_141416002 | 1.40 |
ENSMUST00000006378.2
ENSMUST00000105788.1 |
Clcnkb
|
chloride channel Kb |
| chr11_+_87793722 | 1.35 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr8_+_72761868 | 1.32 |
ENSMUST00000058099.8
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr8_-_71723308 | 1.29 |
ENSMUST00000125092.1
|
Fcho1
|
FCH domain only 1 |
| chr18_+_67343564 | 1.24 |
ENSMUST00000025404.8
|
Cidea
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| chr11_-_102365111 | 1.24 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr8_+_23139064 | 1.23 |
ENSMUST00000033947.8
|
Ank1
|
ankyrin 1, erythroid |
| chr4_-_119189949 | 1.23 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr1_-_170927540 | 1.21 |
ENSMUST00000162136.1
ENSMUST00000162887.1 |
Fcrla
|
Fc receptor-like A |
| chr1_-_170927567 | 1.17 |
ENSMUST00000046322.7
ENSMUST00000159171.1 |
Fcrla
|
Fc receptor-like A |
| chr9_+_45403138 | 1.16 |
ENSMUST00000041005.5
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr10_+_77893413 | 1.15 |
ENSMUST00000178996.1
|
1700009J07Rik
|
RIKEN cDNA 1700009J07 gene |
| chr17_-_35085609 | 1.12 |
ENSMUST00000038507.6
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr14_-_63417125 | 1.10 |
ENSMUST00000014597.3
|
Blk
|
B lymphoid kinase |
| chr13_+_55399648 | 1.08 |
ENSMUST00000057167.7
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr10_+_75564086 | 1.07 |
ENSMUST00000141062.1
ENSMUST00000152657.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_-_37719782 | 1.07 |
ENSMUST00000160589.1
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene |
| chr8_+_23139030 | 1.02 |
ENSMUST00000121075.1
|
Ank1
|
ankyrin 1, erythroid |
| chr14_-_43875517 | 0.98 |
ENSMUST00000179200.1
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr17_+_25298389 | 0.96 |
ENSMUST00000037453.2
|
Prss34
|
protease, serine, 34 |
| chr3_-_92621173 | 0.93 |
ENSMUST00000170676.2
|
Lce6a
|
late cornified envelope 6A |
| chr4_+_115299046 | 0.93 |
ENSMUST00000084343.3
|
Cyp4a12a
|
cytochrome P450, family 4, subfamily a, polypeptide 12a |
| chr11_+_77009265 | 0.92 |
ENSMUST00000129572.2
|
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr11_+_32286946 | 0.89 |
ENSMUST00000101387.3
|
Hbq1b
|
hemoglobin, theta 1B |
| chr11_+_104577281 | 0.87 |
ENSMUST00000106956.3
|
Myl4
|
myosin, light polypeptide 4 |
| chr4_+_46039202 | 0.87 |
ENSMUST00000156200.1
|
Tmod1
|
tropomodulin 1 |
| chr6_+_83743010 | 0.85 |
ENSMUST00000006431.6
|
Atp6v1b1
|
ATPase, H+ transporting, lysosomal V1 subunit B1 |
| chr18_+_36528145 | 0.83 |
ENSMUST00000074298.6
ENSMUST00000115694.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr9_-_42124276 | 0.83 |
ENSMUST00000060989.8
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr7_-_142656018 | 0.81 |
ENSMUST00000178921.1
|
Igf2
|
insulin-like growth factor 2 |
| chr11_+_58917889 | 0.81 |
ENSMUST00000069941.6
|
Btnl10
|
butyrophilin-like 10 |
| chr6_-_69631933 | 0.81 |
ENSMUST00000177697.1
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr4_-_119190005 | 0.81 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr18_+_64254359 | 0.81 |
ENSMUST00000025477.7
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr6_-_113434529 | 0.80 |
ENSMUST00000133348.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr14_-_70635946 | 0.80 |
ENSMUST00000022695.9
|
Dmtn
|
dematin actin binding protein |
| chr2_+_164948219 | 0.80 |
ENSMUST00000017881.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr2_+_25423234 | 0.80 |
ENSMUST00000134259.1
ENSMUST00000100320.4 |
Fut7
|
fucosyltransferase 7 |
| chr6_-_67535783 | 0.79 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr6_-_124733067 | 0.79 |
ENSMUST00000173647.1
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr8_+_84701430 | 0.78 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr9_-_21963568 | 0.76 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr6_-_69400097 | 0.75 |
ENSMUST00000177795.1
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr5_-_148392810 | 0.73 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_-_4191035 | 0.73 |
ENSMUST00000045864.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr6_+_129512538 | 0.72 |
ENSMUST00000032263.5
|
Tmem52b
|
transmembrane protein 52B |
| chr15_-_103252810 | 0.69 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr11_-_59035064 | 0.67 |
ENSMUST00000138587.1
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr11_-_114960417 | 0.66 |
ENSMUST00000092466.5
ENSMUST00000061637.3 |
Cd300c
|
CD300C antigen |
| chr11_+_116531744 | 0.65 |
ENSMUST00000106387.2
ENSMUST00000100201.3 |
Sphk1
|
sphingosine kinase 1 |
| chr7_-_3898120 | 0.64 |
ENSMUST00000070639.7
|
Gm14548
|
predicted gene 14548 |
| chr7_+_123214776 | 0.63 |
ENSMUST00000131933.1
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr3_-_130061553 | 0.62 |
ENSMUST00000168675.1
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr5_+_122209729 | 0.61 |
ENSMUST00000072602.7
ENSMUST00000143560.1 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr2_+_118111876 | 0.60 |
ENSMUST00000039559.8
|
Thbs1
|
thrombospondin 1 |
| chr5_-_104077608 | 0.59 |
ENSMUST00000164471.1
ENSMUST00000178967.1 |
Gm17660
|
predicted gene, 17660 |
| chr4_-_41098174 | 0.59 |
ENSMUST00000055327.7
|
Aqp3
|
aquaporin 3 |
| chr17_+_55970451 | 0.58 |
ENSMUST00000044216.6
|
Shd
|
src homology 2 domain-containing transforming protein D |
| chr4_+_105789869 | 0.57 |
ENSMUST00000184254.1
|
Gm12728
|
predicted gene 12728 |
| chr14_-_51057242 | 0.56 |
ENSMUST00000089798.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr7_+_30422389 | 0.55 |
ENSMUST00000108175.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr4_+_119255494 | 0.55 |
ENSMUST00000094823.3
|
Cldn19
|
claudin 19 |
| chrX_-_135210672 | 0.55 |
ENSMUST00000033783.1
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr2_+_181680284 | 0.54 |
ENSMUST00000103042.3
|
Tcea2
|
transcription elongation factor A (SII), 2 |
| chr7_-_3825641 | 0.54 |
ENSMUST00000094911.4
ENSMUST00000108619.1 ENSMUST00000108620.1 |
Gm15448
|
predicted gene 15448 |
| chr11_-_99155067 | 0.54 |
ENSMUST00000103134.3
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr11_+_113619318 | 0.54 |
ENSMUST00000146390.2
ENSMUST00000106630.1 |
Sstr2
|
somatostatin receptor 2 |
| chr17_+_8924109 | 0.53 |
ENSMUST00000149440.1
|
Pde10a
|
phosphodiesterase 10A |
| chr7_+_30699783 | 0.52 |
ENSMUST00000013227.7
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr12_-_8539545 | 0.52 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr12_+_109549157 | 0.51 |
ENSMUST00000128458.1
ENSMUST00000150851.1 |
Meg3
|
maternally expressed 3 |
| chr2_+_105127200 | 0.51 |
ENSMUST00000139585.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr7_+_30421724 | 0.51 |
ENSMUST00000108176.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr12_+_31073962 | 0.51 |
ENSMUST00000041133.8
|
Fam110c
|
family with sequence similarity 110, member C |
| chr15_+_80623499 | 0.51 |
ENSMUST00000043149.7
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr4_-_49597425 | 0.51 |
ENSMUST00000150664.1
|
Tmem246
|
transmembrane protein 246 |
| chr6_-_5496296 | 0.48 |
ENSMUST00000019721.4
|
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr16_-_20426375 | 0.48 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr4_-_111902754 | 0.47 |
ENSMUST00000102719.1
ENSMUST00000102721.1 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr7_-_29125142 | 0.47 |
ENSMUST00000179893.1
ENSMUST00000032813.9 |
Ryr1
|
ryanodine receptor 1, skeletal muscle |
| chr10_-_24092320 | 0.47 |
ENSMUST00000092654.2
|
Taar8b
|
trace amine-associated receptor 8B |
| chr10_-_81500132 | 0.47 |
ENSMUST00000053646.5
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr6_+_112459501 | 0.47 |
ENSMUST00000075477.6
|
Cav3
|
caveolin 3 |
| chr9_-_110645328 | 0.47 |
ENSMUST00000149089.1
|
Nbeal2
|
neurobeachin-like 2 |
| chr7_-_126704179 | 0.47 |
ENSMUST00000106364.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr5_+_134676490 | 0.47 |
ENSMUST00000100641.2
|
Gm10369
|
predicted gene 10369 |
| chr7_+_28440927 | 0.47 |
ENSMUST00000078845.6
|
Gmfg
|
glia maturation factor, gamma |
| chr6_+_145121727 | 0.47 |
ENSMUST00000032396.6
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr7_+_99535652 | 0.47 |
ENSMUST00000032995.8
ENSMUST00000162404.1 |
Arrb1
|
arrestin, beta 1 |
| chr15_+_80097866 | 0.46 |
ENSMUST00000143928.1
|
Syngr1
|
synaptogyrin 1 |
| chr4_-_156059414 | 0.46 |
ENSMUST00000184348.1
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chrX_-_112405822 | 0.46 |
ENSMUST00000124335.1
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr15_-_102189032 | 0.45 |
ENSMUST00000023805.1
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr3_+_96181151 | 0.45 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr7_+_110777653 | 0.45 |
ENSMUST00000148292.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr11_-_94782500 | 0.45 |
ENSMUST00000162809.2
|
Tmem92
|
transmembrane protein 92 |
| chr5_-_122697603 | 0.45 |
ENSMUST00000071235.4
|
Gm10064
|
predicted gene 10064 |
| chr15_-_82224330 | 0.45 |
ENSMUST00000089161.2
ENSMUST00000109535.2 |
Tnfrsf13c
|
tumor necrosis factor receptor superfamily, member 13c |
| chr16_-_20426322 | 0.44 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr5_+_145345254 | 0.44 |
ENSMUST00000079268.7
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr7_-_141117772 | 0.44 |
ENSMUST00000067836.7
|
Ano9
|
anoctamin 9 |
| chr17_+_35821675 | 0.43 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chrX_+_136666375 | 0.43 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr3_+_10012548 | 0.43 |
ENSMUST00000029046.8
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr5_+_37050854 | 0.43 |
ENSMUST00000043794.4
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr6_-_85374606 | 0.43 |
ENSMUST00000060837.7
|
Rab11fip5
|
RAB11 family interacting protein 5 (class I) |
| chr7_+_28441026 | 0.43 |
ENSMUST00000135686.1
|
Gmfg
|
glia maturation factor, gamma |
| chr2_+_32628390 | 0.41 |
ENSMUST00000156578.1
|
Ak1
|
adenylate kinase 1 |
| chr11_-_69948145 | 0.41 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr9_+_110344185 | 0.41 |
ENSMUST00000142100.1
|
Scap
|
SREBF chaperone |
| chr5_+_77016023 | 0.40 |
ENSMUST00000031161.4
ENSMUST00000117880.1 |
1700023E05Rik
|
RIKEN cDNA 1700023E05 gene |
| chr7_-_3720382 | 0.40 |
ENSMUST00000078451.6
|
Pirb
|
paired Ig-like receptor B |
| chr8_-_35826435 | 0.40 |
ENSMUST00000060128.5
|
Cldn23
|
claudin 23 |
| chr4_+_132974102 | 0.40 |
ENSMUST00000030693.6
|
Fgr
|
Gardner-Rasheed feline sarcoma viral (Fgr) oncogene homolog |
| chr11_+_7063423 | 0.39 |
ENSMUST00000020706.4
|
Adcy1
|
adenylate cyclase 1 |
| chr8_-_70487314 | 0.39 |
ENSMUST00000045286.7
|
Tmem59l
|
transmembrane protein 59-like |
| chr19_+_24673998 | 0.39 |
ENSMUST00000057243.4
|
Tmem252
|
transmembrane protein 252 |
| chr4_-_133874682 | 0.38 |
ENSMUST00000168974.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr9_+_111004811 | 0.38 |
ENSMUST00000080872.4
|
Gm10030
|
predicted gene 10030 |
| chr18_+_34624621 | 0.38 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr14_+_27000362 | 0.38 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr18_-_56925509 | 0.38 |
ENSMUST00000102912.1
|
March3
|
membrane-associated ring finger (C3HC4) 3 |
| chr12_+_69168808 | 0.38 |
ENSMUST00000110621.1
|
Lrr1
|
leucine rich repeat protein 1 |
| chr8_+_84021444 | 0.38 |
ENSMUST00000055077.6
|
Palm3
|
paralemmin 3 |
| chr16_-_19983005 | 0.37 |
ENSMUST00000058839.8
|
Klhl6
|
kelch-like 6 |
| chr4_-_156050465 | 0.37 |
ENSMUST00000184684.1
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr9_+_54764748 | 0.37 |
ENSMUST00000034830.8
|
Crabp1
|
cellular retinoic acid binding protein I |
| chr6_+_119236507 | 0.36 |
ENSMUST00000037434.6
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr9_-_63711969 | 0.36 |
ENSMUST00000154323.1
|
Smad3
|
SMAD family member 3 |
| chr11_-_94782703 | 0.36 |
ENSMUST00000100554.1
|
Tmem92
|
transmembrane protein 92 |
| chr17_+_34092340 | 0.36 |
ENSMUST00000025192.7
|
H2-Oa
|
histocompatibility 2, O region alpha locus |
| chr19_-_5726240 | 0.36 |
ENSMUST00000049295.8
ENSMUST00000075606.4 |
Ehbp1l1
|
EH domain binding protein 1-like 1 |
| chr1_-_175692624 | 0.36 |
ENSMUST00000027809.7
|
Opn3
|
opsin 3 |
| chr12_+_109747903 | 0.35 |
ENSMUST00000183084.1
ENSMUST00000182300.1 |
Mirg
|
miRNA containing gene |
| chr15_+_78899755 | 0.35 |
ENSMUST00000001226.3
ENSMUST00000061239.7 ENSMUST00000109698.2 |
Sh3bp1
|
SH3-domain binding protein 1 |
| chr15_-_95830072 | 0.35 |
ENSMUST00000168960.1
|
Gm17546
|
predicted gene, 17546 |
| chr15_-_74763567 | 0.34 |
ENSMUST00000040404.6
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr18_-_34597468 | 0.34 |
ENSMUST00000056932.2
|
4933408B17Rik
|
RIKEN cDNA 4933408B17 gene |
| chr5_-_136883115 | 0.34 |
ENSMUST00000057497.6
ENSMUST00000111103.1 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr17_-_35697971 | 0.34 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr7_+_123214808 | 0.34 |
ENSMUST00000033035.6
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr12_-_69790660 | 0.34 |
ENSMUST00000021377.4
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr18_+_34625009 | 0.34 |
ENSMUST00000166044.1
|
Kif20a
|
kinesin family member 20A |
| chr2_+_156421083 | 0.34 |
ENSMUST00000125153.2
ENSMUST00000103136.1 ENSMUST00000109577.2 |
Epb4.1l1
|
erythrocyte protein band 4.1-like 1 |
| chr2_-_32083783 | 0.34 |
ENSMUST00000056406.6
|
Fam78a
|
family with sequence similarity 78, member A |
| chr8_+_70863127 | 0.33 |
ENSMUST00000050921.2
|
A230052G05Rik
|
RIKEN cDNA A230052G05 gene |
| chr16_-_32810477 | 0.33 |
ENSMUST00000179384.2
|
Gm933
|
predicted gene 933 |
| chr19_-_41743665 | 0.33 |
ENSMUST00000025993.3
|
Slit1
|
slit homolog 1 (Drosophila) |
| chrX_+_49463926 | 0.33 |
ENSMUST00000130558.1
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_-_43163891 | 0.33 |
ENSMUST00000008280.7
|
Fhl2
|
four and a half LIM domains 2 |
| chr7_-_30880263 | 0.33 |
ENSMUST00000108125.2
|
Cd22
|
CD22 antigen |
| chr2_-_157337424 | 0.32 |
ENSMUST00000109536.1
|
Ghrh
|
growth hormone releasing hormone |
| chr10_+_128908907 | 0.32 |
ENSMUST00000105229.1
|
Cd63
|
CD63 antigen |
| chr14_+_57999305 | 0.32 |
ENSMUST00000180534.1
|
3110083C13Rik
|
RIKEN cDNA 3110083C13 gene |
| chr15_-_36555556 | 0.32 |
ENSMUST00000161202.1
ENSMUST00000013755.5 |
Snx31
|
sorting nexin 31 |
| chr5_-_114658414 | 0.32 |
ENSMUST00000112225.1
ENSMUST00000071968.2 |
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr4_+_134102581 | 0.32 |
ENSMUST00000074690.4
ENSMUST00000070246.2 ENSMUST00000156750.1 |
Ubxn11
|
UBX domain protein 11 |
| chr19_+_53529100 | 0.31 |
ENSMUST00000038287.6
|
Dusp5
|
dual specificity phosphatase 5 |
| chr9_+_46268601 | 0.31 |
ENSMUST00000121598.1
|
Apoa5
|
apolipoprotein A-V |
| chr1_+_134962553 | 0.31 |
ENSMUST00000027687.7
|
Ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr1_+_131019843 | 0.31 |
ENSMUST00000016673.5
|
Il10
|
interleukin 10 |
| chr18_+_44104407 | 0.31 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr7_-_3249711 | 0.31 |
ENSMUST00000108653.2
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr7_+_46847128 | 0.31 |
ENSMUST00000005051.4
|
Ldha
|
lactate dehydrogenase A |
| chr14_+_63046988 | 0.30 |
ENSMUST00000067990.1
ENSMUST00000111203.1 |
Defb42
|
defensin beta 42 |
| chr12_-_84698769 | 0.30 |
ENSMUST00000095550.2
|
Syndig1l
|
synapse differentiation inducing 1 like |
| chr6_-_112388013 | 0.30 |
ENSMUST00000060847.5
|
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr11_+_87760533 | 0.30 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr5_+_114146525 | 0.30 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr11_-_100356078 | 0.29 |
ENSMUST00000103124.4
|
Hap1
|
huntingtin-associated protein 1 |
| chr13_-_41487306 | 0.29 |
ENSMUST00000021794.6
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr10_+_81176631 | 0.29 |
ENSMUST00000047864.9
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chrX_-_112406779 | 0.29 |
ENSMUST00000026601.2
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr1_+_152807877 | 0.29 |
ENSMUST00000027754.6
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr4_-_132345715 | 0.29 |
ENSMUST00000084250.4
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr1_+_191063001 | 0.29 |
ENSMUST00000076952.5
ENSMUST00000139340.1 ENSMUST00000078259.6 |
Nsl1
|
NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr15_-_82794236 | 0.29 |
ENSMUST00000006094.4
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr6_+_66535390 | 0.28 |
ENSMUST00000116605.1
|
Mad2l1
|
MAD2 mitotic arrest deficient-like 1 |
| chr14_-_98169542 | 0.28 |
ENSMUST00000069334.7
ENSMUST00000071533.6 |
Dach1
|
dachshund 1 (Drosophila) |
| chr6_-_129717132 | 0.28 |
ENSMUST00000088046.1
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr9_-_97111117 | 0.28 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr10_+_83722865 | 0.28 |
ENSMUST00000150459.1
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr2_+_164698501 | 0.28 |
ENSMUST00000017454.7
|
Spint4
|
serine protease inhibitor, Kunitz type 4 |
| chr6_-_135254326 | 0.28 |
ENSMUST00000111911.2
ENSMUST00000111910.2 |
Gsg1
|
germ cell-specific gene 1 |
| chr7_+_19282613 | 0.28 |
ENSMUST00000032559.9
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr11_-_69822144 | 0.28 |
ENSMUST00000045771.6
|
Spem1
|
sperm maturation 1 |
| chr3_-_59210881 | 0.28 |
ENSMUST00000040622.1
|
P2ry13
|
purinergic receptor P2Y, G-protein coupled 13 |
| chr11_+_105994635 | 0.28 |
ENSMUST00000183675.1
ENSMUST00000184871.1 |
ACE3
Ace3
|
angiotensin I converting enzyme 3 precursor angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.5 | 1.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) neutrophil mediated killing of fungus(GO:0070947) |
| 0.4 | 1.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 0.9 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.3 | 1.7 | GO:1902732 | antifungal humoral response(GO:0019732) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.3 | 0.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.3 | 0.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 3.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 0.9 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.2 | 1.8 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.2 | 0.8 | GO:2001137 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 0.6 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 | 1.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.2 | 0.9 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 0.6 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.2 | 0.5 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.2 | 0.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 0.3 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.2 | 0.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 1.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.5 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.7 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.6 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.1 | 0.6 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.4 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.4 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.1 | 0.4 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.6 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 0.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.4 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.1 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) positive regulation of thymocyte apoptotic process(GO:0070245) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.1 | 0.3 | GO:0002874 | negative regulation of cytokine secretion involved in immune response(GO:0002740) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) negative regulation of sensory perception of pain(GO:1904057) |
| 0.1 | 1.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.1 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.1 | 0.4 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.5 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 0.3 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.3 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 0.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.2 | GO:0071725 | detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 1.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.1 | 0.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.1 | 0.8 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.2 | GO:0033624 | negative regulation of integrin activation(GO:0033624) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.5 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.5 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.2 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 2.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.3 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.5 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.5 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 1.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.5 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.5 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.2 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) negative regulation of primary amine oxidase activity(GO:1902283) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.3 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.2 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 1.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 1.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0070572 | positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.4 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 1.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.8 | GO:0045851 | pH reduction(GO:0045851) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 2.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.4 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.0 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.0 | 0.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.0 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 4.8 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 0.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.0 | 0.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.1 | GO:1990452 | LUBAC complex(GO:0071797) Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 0.9 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.3 | 0.8 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.2 | 2.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 0.7 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 1.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 0.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 0.9 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.2 | 0.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.6 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 3.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.6 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.7 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.2 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 4.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) lipopeptide binding(GO:0071723) |
| 0.1 | 0.2 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0070401 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.0 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 0.0 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 4.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 2.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.7 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 1.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |