Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
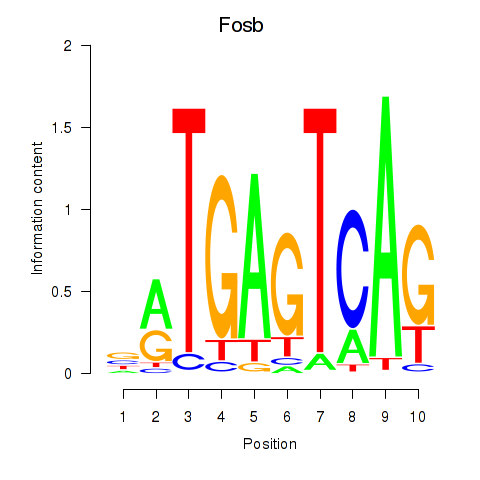
Results for Fosb
Z-value: 0.58
Transcription factors associated with Fosb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosb
|
ENSMUSG00000003545.2 | FBJ osteosarcoma oncogene B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosb | mm10_v2_chr7_-_19310035_19310050 | -0.35 | 3.5e-02 | Click! |
Activity profile of Fosb motif
Sorted Z-values of Fosb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_7445822 | 1.34 |
ENSMUST00000034497.6
|
Mmp3
|
matrix metallopeptidase 3 |
| chr5_-_24392012 | 0.82 |
ENSMUST00000059401.6
|
Atg9b
|
autophagy related 9B |
| chr1_+_165769392 | 0.61 |
ENSMUST00000040298.4
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr17_-_45592485 | 0.52 |
ENSMUST00000166119.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_+_126776939 | 0.49 |
ENSMUST00000038614.5
ENSMUST00000170882.1 ENSMUST00000106359.1 ENSMUST00000106357.1 ENSMUST00000145762.1 ENSMUST00000132643.1 ENSMUST00000106356.1 |
Ypel3
|
yippee-like 3 (Drosophila) |
| chr17_-_45592262 | 0.48 |
ENSMUST00000164769.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr19_+_5877794 | 0.48 |
ENSMUST00000145200.1
ENSMUST00000025732.7 ENSMUST00000125114.1 ENSMUST00000155697.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr10_+_88091070 | 0.46 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr9_+_108953578 | 0.45 |
ENSMUST00000112070.1
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr9_+_108953597 | 0.45 |
ENSMUST00000026740.5
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr11_-_100248902 | 0.44 |
ENSMUST00000007280.8
|
Krt16
|
keratin 16 |
| chr7_+_30787897 | 0.42 |
ENSMUST00000098559.1
|
Krtdap
|
keratinocyte differentiation associated protein |
| chr11_-_120630516 | 0.41 |
ENSMUST00000106181.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr1_-_74893109 | 0.40 |
ENSMUST00000006721.2
|
Cryba2
|
crystallin, beta A2 |
| chrX_-_75578188 | 0.39 |
ENSMUST00000033545.5
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr18_+_44071390 | 0.39 |
ENSMUST00000068473.2
|
Spink6
|
serine peptidase inhibitor, Kazal type 6 |
| chr8_-_110039330 | 0.38 |
ENSMUST00000109222.2
|
Chst4
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 4 |
| chr7_-_4778141 | 0.38 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr11_-_84067063 | 0.37 |
ENSMUST00000108101.1
|
Dusp14
|
dual specificity phosphatase 14 |
| chr10_+_24447468 | 0.36 |
ENSMUST00000134627.1
|
Gm15271
|
predicted gene 15271 |
| chr7_-_133782721 | 0.34 |
ENSMUST00000063669.1
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr3_-_88503331 | 0.34 |
ENSMUST00000029699.6
|
Lmna
|
lamin A |
| chr3_-_88503187 | 0.34 |
ENSMUST00000120377.1
|
Lmna
|
lamin A |
| chr1_-_51915968 | 0.33 |
ENSMUST00000046390.7
|
Myo1b
|
myosin IB |
| chr12_-_85288419 | 0.33 |
ENSMUST00000121930.1
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr11_-_69549108 | 0.33 |
ENSMUST00000108659.1
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr7_+_141079125 | 0.33 |
ENSMUST00000159375.1
|
Pkp3
|
plakophilin 3 |
| chr4_+_42158092 | 0.33 |
ENSMUST00000098122.2
|
Gm13306
|
predicted gene 13306 |
| chr5_+_31116702 | 0.33 |
ENSMUST00000013771.8
|
Trim54
|
tripartite motif-containing 54 |
| chr17_-_30612613 | 0.31 |
ENSMUST00000167624.1
ENSMUST00000024823.6 |
Glo1
|
glyoxalase 1 |
| chr9_+_44240668 | 0.29 |
ENSMUST00000092426.3
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr11_+_80428598 | 0.28 |
ENSMUST00000173938.1
ENSMUST00000017572.7 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr7_-_80401707 | 0.28 |
ENSMUST00000120753.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_-_4139605 | 0.28 |
ENSMUST00000025761.6
|
Cabp4
|
calcium binding protein 4 |
| chr5_+_31116722 | 0.27 |
ENSMUST00000114637.1
|
Trim54
|
tripartite motif-containing 54 |
| chr13_-_54611274 | 0.26 |
ENSMUST00000049575.7
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr10_-_86011833 | 0.25 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr10_+_26078255 | 0.25 |
ENSMUST00000041011.3
|
Gm9767
|
predicted gene 9767 |
| chr14_+_66635251 | 0.24 |
ENSMUST00000159365.1
ENSMUST00000054661.1 ENSMUST00000159068.1 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr13_-_54611332 | 0.23 |
ENSMUST00000091609.4
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr2_+_143915273 | 0.23 |
ENSMUST00000103172.3
|
Dstn
|
destrin |
| chr2_-_151632471 | 0.23 |
ENSMUST00000137936.1
ENSMUST00000146172.1 ENSMUST00000094456.3 ENSMUST00000148755.1 ENSMUST00000109875.1 ENSMUST00000028951.7 ENSMUST00000109877.3 |
Snph
|
syntaphilin |
| chr7_+_139834148 | 0.23 |
ENSMUST00000026548.7
|
Gpr123
|
G protein-coupled receptor 123 |
| chr2_+_30441831 | 0.22 |
ENSMUST00000131476.1
|
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr9_+_7272514 | 0.22 |
ENSMUST00000015394.8
|
Mmp13
|
matrix metallopeptidase 13 |
| chr7_-_4971168 | 0.21 |
ENSMUST00000133272.1
|
Gm1078
|
predicted gene 1078 |
| chr5_-_107875035 | 0.20 |
ENSMUST00000138111.1
ENSMUST00000112642.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr9_-_119977250 | 0.19 |
ENSMUST00000035101.7
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr16_+_17561885 | 0.18 |
ENSMUST00000171002.1
ENSMUST00000023441.4 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr6_+_29529277 | 0.18 |
ENSMUST00000163511.1
|
Irf5
|
interferon regulatory factor 5 |
| chr1_-_10719898 | 0.17 |
ENSMUST00000035577.6
|
Cpa6
|
carboxypeptidase A6 |
| chr9_+_104566677 | 0.17 |
ENSMUST00000157006.1
|
Cpne4
|
copine IV |
| chr16_+_20548577 | 0.17 |
ENSMUST00000003319.5
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr7_+_30763750 | 0.17 |
ENSMUST00000165887.1
ENSMUST00000085691.4 ENSMUST00000085688.4 ENSMUST00000054427.6 |
Dmkn
|
dermokine |
| chr10_-_64090241 | 0.17 |
ENSMUST00000133588.1
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr2_-_18048784 | 0.16 |
ENSMUST00000142856.1
|
Skida1
|
SKI/DACH domain containing 1 |
| chr18_+_40258361 | 0.16 |
ENSMUST00000091927.4
|
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr12_-_84194007 | 0.16 |
ENSMUST00000110294.1
|
Elmsan1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr9_+_7347374 | 0.16 |
ENSMUST00000065079.5
ENSMUST00000005950.5 |
Mmp12
|
matrix metallopeptidase 12 |
| chr1_-_171240055 | 0.16 |
ENSMUST00000131286.1
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr18_-_31317043 | 0.16 |
ENSMUST00000139924.1
ENSMUST00000153060.1 |
Rit2
|
Ras-like without CAAX 2 |
| chr9_-_109059216 | 0.16 |
ENSMUST00000112053.1
|
Trex1
|
three prime repair exonuclease 1 |
| chr1_+_134560157 | 0.15 |
ENSMUST00000047714.7
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr7_-_127946725 | 0.15 |
ENSMUST00000118755.1
ENSMUST00000094026.3 |
Prss36
|
protease, serine, 36 |
| chr1_-_12991109 | 0.14 |
ENSMUST00000115403.2
ENSMUST00000115402.1 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr18_-_43477764 | 0.14 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr6_-_124769548 | 0.14 |
ENSMUST00000149652.1
ENSMUST00000112476.1 ENSMUST00000004378.8 |
Eno2
|
enolase 2, gamma neuronal |
| chr11_-_50210765 | 0.13 |
ENSMUST00000143379.1
ENSMUST00000015981.5 ENSMUST00000102774.4 |
Sqstm1
|
sequestosome 1 |
| chr11_+_61684419 | 0.13 |
ENSMUST00000093019.5
|
Fam83g
|
family with sequence similarity 83, member G |
| chr17_-_42742167 | 0.12 |
ENSMUST00000113614.2
|
Gpr111
|
G protein-coupled receptor 111 |
| chr11_-_3914664 | 0.12 |
ENSMUST00000109995.1
ENSMUST00000051207.1 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr4_-_19922599 | 0.12 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr3_-_123034943 | 0.12 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr10_+_116143881 | 0.12 |
ENSMUST00000105271.2
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr7_+_26435614 | 0.12 |
ENSMUST00000119386.1
|
Nlrp4a
|
NLR family, pyrin domain containing 4A |
| chr17_+_47769191 | 0.12 |
ENSMUST00000160373.1
ENSMUST00000159641.1 |
Tfeb
|
transcription factor EB |
| chr10_-_64090265 | 0.11 |
ENSMUST00000105439.1
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_-_88177671 | 0.10 |
ENSMUST00000181837.1
|
1700113A16Rik
|
RIKEN cDNA 1700113A16 gene |
| chr9_-_109059711 | 0.10 |
ENSMUST00000061973.4
|
Trex1
|
three prime repair exonuclease 1 |
| chr16_+_32400506 | 0.10 |
ENSMUST00000115149.2
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chr11_-_96075655 | 0.10 |
ENSMUST00000090541.5
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr15_-_76014318 | 0.10 |
ENSMUST00000060807.5
|
Fam83h
|
family with sequence similarity 83, member H |
| chr12_+_71015966 | 0.10 |
ENSMUST00000046305.5
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr11_+_77801325 | 0.10 |
ENSMUST00000151373.2
ENSMUST00000172303.2 ENSMUST00000130305.2 ENSMUST00000164334.1 ENSMUST00000092884.4 |
Myo18a
|
myosin XVIIIA |
| chr11_+_77801291 | 0.09 |
ENSMUST00000100794.3
|
Myo18a
|
myosin XVIIIA |
| chr14_-_55758458 | 0.09 |
ENSMUST00000001497.7
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr12_+_51690966 | 0.09 |
ENSMUST00000021338.8
|
Ap4s1
|
adaptor-related protein complex AP-4, sigma 1 |
| chr9_+_7464141 | 0.09 |
ENSMUST00000034492.5
|
Mmp1a
|
matrix metallopeptidase 1a (interstitial collagenase) |
| chr11_-_96075581 | 0.09 |
ENSMUST00000107686.1
ENSMUST00000107684.1 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr3_+_116878227 | 0.09 |
ENSMUST00000040260.6
|
Frrs1
|
ferric-chelate reductase 1 |
| chr15_+_102503722 | 0.09 |
ENSMUST00000096145.4
|
Gm10337
|
predicted gene 10337 |
| chr2_+_110721587 | 0.09 |
ENSMUST00000111017.2
|
Muc15
|
mucin 15 |
| chr8_+_4226827 | 0.08 |
ENSMUST00000053035.6
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr7_-_105787567 | 0.07 |
ENSMUST00000144189.1
|
Dchs1
|
dachsous 1 (Drosophila) |
| chr2_-_25224653 | 0.07 |
ENSMUST00000043584.4
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr2_+_110721340 | 0.07 |
ENSMUST00000111016.2
|
Muc15
|
mucin 15 |
| chr7_+_30776394 | 0.06 |
ENSMUST00000041703.7
|
Dmkn
|
dermokine |
| chr11_-_109473598 | 0.06 |
ENSMUST00000070152.5
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr3_+_123267445 | 0.06 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr10_-_44458715 | 0.06 |
ENSMUST00000039174.4
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr7_+_6045161 | 0.06 |
ENSMUST00000037728.5
ENSMUST00000121583.1 |
Nlrp4c
|
NLR family, pyrin domain containing 4C |
| chr15_+_101473472 | 0.06 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr1_+_106861144 | 0.06 |
ENSMUST00000086701.6
ENSMUST00000112730.1 |
Serpinb5
|
serine (or cysteine) peptidase inhibitor, clade B, member 5 |
| chr1_+_120602405 | 0.06 |
ENSMUST00000079721.7
|
En1
|
engrailed 1 |
| chr3_+_55461758 | 0.06 |
ENSMUST00000070418.4
|
Dclk1
|
doublecortin-like kinase 1 |
| chr18_+_42053692 | 0.06 |
ENSMUST00000074679.3
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr13_-_113663670 | 0.05 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr11_+_4257557 | 0.05 |
ENSMUST00000066283.5
|
Lif
|
leukemia inhibitory factor |
| chr1_+_134560190 | 0.05 |
ENSMUST00000112198.1
ENSMUST00000112197.1 |
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr9_+_96259246 | 0.04 |
ENSMUST00000179065.1
ENSMUST00000165768.2 |
Tfdp2
|
transcription factor Dp 2 |
| chr2_+_118663235 | 0.04 |
ENSMUST00000099557.3
|
Pak6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr9_+_96258697 | 0.04 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr4_-_140665891 | 0.04 |
ENSMUST00000069623.5
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr6_+_42349826 | 0.03 |
ENSMUST00000070635.6
|
Zyx
|
zyxin |
| chr1_+_182564994 | 0.03 |
ENSMUST00000048941.7
ENSMUST00000168514.1 |
Capn8
|
calpain 8 |
| chr11_+_117232254 | 0.03 |
ENSMUST00000106354.2
|
Sept9
|
septin 9 |
| chr17_+_78508063 | 0.03 |
ENSMUST00000024880.9
|
Vit
|
vitrin |
| chr14_+_27622433 | 0.03 |
ENSMUST00000090302.5
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr2_+_151572606 | 0.03 |
ENSMUST00000028950.8
|
Sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr19_-_38125258 | 0.02 |
ENSMUST00000025951.6
|
Rbp4
|
retinol binding protein 4, plasma |
| chr12_+_83632208 | 0.02 |
ENSMUST00000048155.9
ENSMUST00000182618.1 ENSMUST00000183154.1 ENSMUST00000182036.1 ENSMUST00000182347.1 |
Rbm25
|
RNA binding motif protein 25 |
| chr16_-_4880284 | 0.01 |
ENSMUST00000037843.6
|
Ubald1
|
UBA-like domain containing 1 |
| chr13_+_30659999 | 0.01 |
ENSMUST00000091672.6
ENSMUST00000110310.1 ENSMUST00000095914.5 |
Dusp22
|
dual specificity phosphatase 22 |
| chr6_+_42350000 | 0.01 |
ENSMUST00000164375.1
|
Zyx
|
zyxin |
| chr2_+_152105722 | 0.00 |
ENSMUST00000099225.2
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 1.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.2 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.2 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.4 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |