Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Foxm1
Z-value: 0.77
Transcription factors associated with Foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxm1
|
ENSMUSG00000001517.8 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxm1 | mm10_v2_chr6_+_128362919_128363058 | -0.19 | 2.7e-01 | Click! |
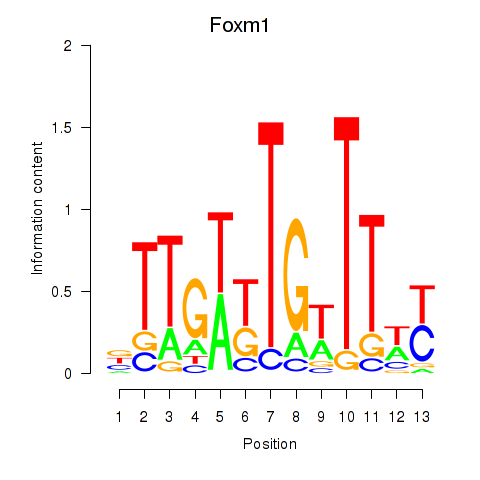
Activity profile of Foxm1 motif
Sorted Z-values of Foxm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_58759700 | 3.70 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr16_-_23890805 | 2.18 |
ENSMUST00000004480.3
|
Sst
|
somatostatin |
| chr17_-_31144271 | 1.79 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr1_-_79440039 | 1.70 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chr10_-_88605017 | 1.61 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr19_+_39992424 | 1.44 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr8_-_41215146 | 1.09 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr11_+_104550663 | 1.06 |
ENSMUST00000018800.2
|
Myl4
|
myosin, light polypeptide 4 |
| chr4_+_44943727 | 1.01 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr6_+_122513583 | 0.84 |
ENSMUST00000032210.7
ENSMUST00000148517.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr9_+_22454290 | 0.73 |
ENSMUST00000168332.1
|
Gm17545
|
predicted gene, 17545 |
| chr10_+_23851454 | 0.61 |
ENSMUST00000020190.7
|
Vnn3
|
vanin 3 |
| chr18_-_43737186 | 0.60 |
ENSMUST00000025381.2
|
Spink3
|
serine peptidase inhibitor, Kazal type 3 |
| chr3_+_138217814 | 0.48 |
ENSMUST00000090171.5
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_+_130865669 | 0.48 |
ENSMUST00000038829.5
|
Faim3
|
Fas apoptotic inhibitory molecule 3 |
| chr7_+_45215753 | 0.47 |
ENSMUST00000033060.6
ENSMUST00000155313.1 ENSMUST00000107801.1 |
Tead2
|
TEA domain family member 2 |
| chr2_-_32424005 | 0.46 |
ENSMUST00000113307.2
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr6_+_37530204 | 0.45 |
ENSMUST00000151256.1
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr8_-_65129317 | 0.45 |
ENSMUST00000098713.3
|
BC030870
|
cDNA sequence BC030870 |
| chrX_-_75578188 | 0.42 |
ENSMUST00000033545.5
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr13_-_12520377 | 0.42 |
ENSMUST00000179308.1
|
Edaradd
|
EDAR (ectodysplasin-A receptor)-associated death domain |
| chr19_-_58455903 | 0.41 |
ENSMUST00000131877.1
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr4_+_107707056 | 0.41 |
ENSMUST00000147654.1
|
1700047F07Rik
|
RIKEN cDNA 1700047F07 gene |
| chr10_+_128267997 | 0.40 |
ENSMUST00000050901.2
|
Apof
|
apolipoprotein F |
| chr1_+_74713551 | 0.39 |
ENSMUST00000027356.5
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr17_+_34204080 | 0.35 |
ENSMUST00000138491.1
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr9_-_108263706 | 0.35 |
ENSMUST00000171412.1
|
Dag1
|
dystroglycan 1 |
| chr4_-_84546284 | 0.35 |
ENSMUST00000177040.1
|
Bnc2
|
basonuclin 2 |
| chr4_-_114908892 | 0.34 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr10_+_127849917 | 0.34 |
ENSMUST00000077530.2
|
Rdh19
|
retinol dehydrogenase 19 |
| chr9_+_85312773 | 0.33 |
ENSMUST00000113205.1
|
Gm11114
|
predicted gene 11114 |
| chr3_-_17230976 | 0.32 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr9_-_71163224 | 0.32 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr19_+_6184363 | 0.32 |
ENSMUST00000025699.2
ENSMUST00000113528.1 |
1700123I01Rik
|
RIKEN cDNA 1700123I01 gene |
| chr7_+_15882617 | 0.31 |
ENSMUST00000125993.1
ENSMUST00000130566.1 |
Crxos1
|
Crx opposite strand transcript 1 |
| chr2_+_68117713 | 0.30 |
ENSMUST00000112346.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr3_+_19985612 | 0.30 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr2_+_136052180 | 0.29 |
ENSMUST00000144403.1
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr1_+_74125535 | 0.28 |
ENSMUST00000080167.4
ENSMUST00000127134.1 |
Rufy4
|
RUN and FYVE domain containing 4 |
| chr6_+_37530173 | 0.28 |
ENSMUST00000040987.7
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr10_-_20725023 | 0.27 |
ENSMUST00000020165.7
|
Pde7b
|
phosphodiesterase 7B |
| chr3_+_81999461 | 0.27 |
ENSMUST00000107736.1
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chrX_+_59999436 | 0.27 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr10_+_96616998 | 0.26 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr11_-_100354040 | 0.26 |
ENSMUST00000173630.1
|
Hap1
|
huntingtin-associated protein 1 |
| chr2_+_135659625 | 0.25 |
ENSMUST00000134310.1
|
Plcb4
|
phospholipase C, beta 4 |
| chr5_+_137553517 | 0.23 |
ENSMUST00000136088.1
ENSMUST00000139395.1 ENSMUST00000136565.1 ENSMUST00000149292.1 ENSMUST00000125489.1 |
Actl6b
|
actin-like 6B |
| chr17_+_28530834 | 0.23 |
ENSMUST00000025060.2
|
Armc12
|
armadillo repeat containing 12 |
| chr17_+_34238896 | 0.23 |
ENSMUST00000095342.3
|
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr3_-_106167564 | 0.23 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chr6_+_48537560 | 0.22 |
ENSMUST00000040361.5
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr7_-_102857271 | 0.21 |
ENSMUST00000071393.2
|
Olfr566
|
olfactory receptor 566 |
| chr9_-_100571049 | 0.21 |
ENSMUST00000093792.2
|
Slc35g2
|
solute carrier family 35, member G2 |
| chr17_-_23645264 | 0.21 |
ENSMUST00000024696.7
|
Mmp25
|
matrix metallopeptidase 25 |
| chr1_-_174118014 | 0.20 |
ENSMUST00000063030.3
|
Olfr231
|
olfactory receptor 231 |
| chrY_-_9132561 | 0.20 |
ENSMUST00000171947.2
|
Gm21292
|
predicted gene, 21292 |
| chr4_+_143413002 | 0.20 |
ENSMUST00000155157.1
|
Pramef8
|
PRAME family member 8 |
| chr19_-_58455161 | 0.19 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_-_39649046 | 0.19 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr14_+_26082245 | 0.19 |
ENSMUST00000168149.2
ENSMUST00000163305.1 |
Cphx2
Cphx3
|
cytoplasmic polyadenylated homeobox 2 cytoplasmic polyadenylated homeobox 3 |
| chr4_+_143412920 | 0.19 |
ENSMUST00000132915.1
ENSMUST00000037356.7 |
Pramef8
|
PRAME family member 8 |
| chr7_-_107625161 | 0.19 |
ENSMUST00000060348.2
|
5330417H12Rik
|
RIKEN cDNA 5330417H12 gene |
| chr4_-_44710408 | 0.19 |
ENSMUST00000134968.2
ENSMUST00000173821.1 ENSMUST00000174319.1 ENSMUST00000173733.1 ENSMUST00000172866.1 ENSMUST00000165417.2 ENSMUST00000107825.2 ENSMUST00000102932.3 ENSMUST00000107827.2 ENSMUST00000107826.2 |
Pax5
|
paired box gene 5 |
| chr1_-_86388162 | 0.19 |
ENSMUST00000027440.3
|
Nmur1
|
neuromedin U receptor 1 |
| chr7_+_31059342 | 0.18 |
ENSMUST00000039775.7
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr12_-_87644467 | 0.18 |
ENSMUST00000180181.1
|
Gm16381
|
predicted gene 16381 |
| chr16_-_46155077 | 0.18 |
ENSMUST00000059524.5
|
Gm4737
|
predicted gene 4737 |
| chr19_-_8218832 | 0.17 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr11_-_69617879 | 0.17 |
ENSMUST00000005334.2
|
Shbg
|
sex hormone binding globulin |
| chr18_+_37320374 | 0.17 |
ENSMUST00000078271.2
|
Pcdhb5
|
protocadherin beta 5 |
| chrX_-_103623648 | 0.17 |
ENSMUST00000156211.1
|
Ftx
|
Ftx transcript, Xist regulator (non-protein coding) |
| chr12_-_80760541 | 0.16 |
ENSMUST00000073251.6
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr7_+_45554893 | 0.16 |
ENSMUST00000107752.3
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr2_-_151476153 | 0.15 |
ENSMUST00000080132.2
|
4921509C19Rik
|
RIKEN cDNA 4921509C19 gene |
| chr6_-_59024340 | 0.15 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr2_+_96318014 | 0.15 |
ENSMUST00000135431.1
ENSMUST00000162807.2 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr3_+_33799791 | 0.15 |
ENSMUST00000099153.3
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr2_-_30048827 | 0.15 |
ENSMUST00000113711.2
|
Wdr34
|
WD repeat domain 34 |
| chr14_+_26221856 | 0.15 |
ENSMUST00000184577.1
|
Cphx3
|
cytoplasmic polyadenylated homeobox 3 |
| chr14_+_25942492 | 0.15 |
ENSMUST00000184016.1
|
Cphx1
|
cytoplasmic polyadenylated homeobox 1 |
| chr11_-_97996171 | 0.14 |
ENSMUST00000042971.9
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr4_-_120951664 | 0.14 |
ENSMUST00000106280.1
|
Zfp69
|
zinc finger protein 69 |
| chr18_+_37484955 | 0.14 |
ENSMUST00000053856.4
|
Pcdhb17
|
protocadherin beta 17 |
| chr8_-_24596960 | 0.14 |
ENSMUST00000033956.6
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr19_+_25672408 | 0.13 |
ENSMUST00000053068.5
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chrX_-_103623704 | 0.13 |
ENSMUST00000130063.1
ENSMUST00000125419.1 |
Ftx
|
Ftx transcript, Xist regulator (non-protein coding) |
| chr10_+_112083345 | 0.13 |
ENSMUST00000148897.1
ENSMUST00000020434.3 |
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr18_-_65939048 | 0.13 |
ENSMUST00000025396.3
|
Rax
|
retina and anterior neural fold homeobox |
| chr19_-_4121536 | 0.13 |
ENSMUST00000025767.7
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr6_+_51544513 | 0.12 |
ENSMUST00000179365.1
ENSMUST00000114439.1 |
Snx10
|
sorting nexin 10 |
| chr1_-_153844130 | 0.12 |
ENSMUST00000124558.3
|
Rgsl1
|
regulator of G-protein signaling like 1 |
| chrX_+_7909542 | 0.12 |
ENSMUST00000086274.2
|
Gm10490
|
predicted gene 10490 |
| chr10_-_81230773 | 0.12 |
ENSMUST00000047408.4
|
Atcay
|
ataxia, cerebellar, Cayman type homolog (human) |
| chr11_-_97995863 | 0.11 |
ENSMUST00000107563.1
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr15_+_98167806 | 0.11 |
ENSMUST00000031914.4
|
AI836003
|
expressed sequence AI836003 |
| chr11_+_61653259 | 0.11 |
ENSMUST00000004959.2
|
Grap
|
GRB2-related adaptor protein |
| chr3_-_108889990 | 0.11 |
ENSMUST00000053065.4
ENSMUST00000102620.3 |
Fndc7
|
fibronectin type III domain containing 7 |
| chrX_-_141725181 | 0.11 |
ENSMUST00000067841.7
|
Irs4
|
insulin receptor substrate 4 |
| chr11_-_96075581 | 0.10 |
ENSMUST00000107686.1
ENSMUST00000107684.1 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr17_+_43389436 | 0.10 |
ENSMUST00000113599.1
|
Gpr116
|
G protein-coupled receptor 116 |
| chrX_-_7041619 | 0.10 |
ENSMUST00000115752.1
|
Ccnb3
|
cyclin B3 |
| chr2_-_72813665 | 0.10 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr10_+_112928501 | 0.10 |
ENSMUST00000180464.1
|
Gm26596
|
predicted gene, 26596 |
| chr7_-_126975400 | 0.10 |
ENSMUST00000180704.1
|
D830044I16Rik
|
RIKEN cDNA D830044I16 gene |
| chrX_+_82948861 | 0.10 |
ENSMUST00000114000.1
|
Dmd
|
dystrophin, muscular dystrophy |
| chr7_-_83550258 | 0.10 |
ENSMUST00000177929.1
|
Gm10610
|
predicted gene 10610 |
| chr4_-_117891994 | 0.10 |
ENSMUST00000030265.3
|
Dph2
|
DPH2 homolog (S. cerevisiae) |
| chr7_-_126566364 | 0.09 |
ENSMUST00000032992.5
|
Eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr4_-_144408397 | 0.09 |
ENSMUST00000123854.1
ENSMUST00000030326.3 |
Pramef12
|
PRAME family member 12 |
| chr4_-_143299498 | 0.09 |
ENSMUST00000030317.7
|
Pdpn
|
podoplanin |
| chr5_-_140321524 | 0.09 |
ENSMUST00000031534.6
|
Mad1l1
|
MAD1 mitotic arrest deficient 1-like 1 |
| chr7_-_103741322 | 0.09 |
ENSMUST00000051346.2
|
Olfr629
|
olfactory receptor 629 |
| chr9_-_121839460 | 0.09 |
ENSMUST00000135986.2
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr9_+_109931458 | 0.08 |
ENSMUST00000072772.5
ENSMUST00000035055.8 |
Map4
|
microtubule-associated protein 4 |
| chr19_-_58455398 | 0.08 |
ENSMUST00000026076.7
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr2_+_10372426 | 0.08 |
ENSMUST00000114864.2
ENSMUST00000116594.2 ENSMUST00000041105.6 |
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr2_-_73452666 | 0.08 |
ENSMUST00000151939.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr12_+_101404673 | 0.08 |
ENSMUST00000055156.4
|
Catsperb
|
catsper channel auxiliary subunit beta |
| chr7_-_133123409 | 0.08 |
ENSMUST00000170459.1
ENSMUST00000166400.1 |
Ctbp2
|
C-terminal binding protein 2 |
| chr5_-_73632421 | 0.08 |
ENSMUST00000087177.2
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr1_+_89454769 | 0.07 |
ENSMUST00000027521.8
ENSMUST00000074945.5 |
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chrY_+_21242966 | 0.07 |
ENSMUST00000179095.1
|
Gm20865
|
predicted gene, 20865 |
| chr11_-_23519181 | 0.07 |
ENSMUST00000020527.5
|
1700093K21Rik
|
RIKEN cDNA 1700093K21 gene |
| chrY_+_35569461 | 0.07 |
ENSMUST00000180002.1
|
Gm21872
|
predicted gene, 21872 |
| chr11_-_99422252 | 0.07 |
ENSMUST00000017741.3
|
Krt12
|
keratin 12 |
| chr4_+_107707030 | 0.07 |
ENSMUST00000140321.1
|
1700047F07Rik
|
RIKEN cDNA 1700047F07 gene |
| chrY_-_85529518 | 0.07 |
ENSMUST00000178889.1
|
Gm20854
|
predicted gene, 20854 |
| chrY_-_79149787 | 0.07 |
ENSMUST00000179922.1
|
Gm20917
|
predicted gene, 20917 |
| chr10_+_94576254 | 0.06 |
ENSMUST00000117929.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr14_+_75955003 | 0.06 |
ENSMUST00000050120.2
|
Kctd4
|
potassium channel tetramerisation domain containing 4 |
| chrY_+_25995644 | 0.06 |
ENSMUST00000179070.1
|
Gm21443
|
predicted gene, 21443 |
| chrX_+_112495266 | 0.06 |
ENSMUST00000026602.2
ENSMUST00000113412.2 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr5_-_31526693 | 0.06 |
ENSMUST00000118874.1
ENSMUST00000117642.1 ENSMUST00000065388.4 |
Supt7l
|
suppressor of Ty 7-like |
| chr2_+_136532266 | 0.06 |
ENSMUST00000121717.1
|
Ankef1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr10_+_127421208 | 0.06 |
ENSMUST00000168780.1
|
R3hdm2
|
R3H domain containing 2 |
| chrY_-_82020055 | 0.06 |
ENSMUST00000177769.1
|
Gm21790
|
predicted gene, 21790 |
| chr10_-_63927434 | 0.06 |
ENSMUST00000079279.3
|
Gm10118
|
predicted gene 10118 |
| chr7_-_80901220 | 0.05 |
ENSMUST00000146402.1
ENSMUST00000026816.8 |
Wdr73
|
WD repeat domain 73 |
| chr2_+_112284561 | 0.05 |
ENSMUST00000053666.7
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr16_-_32165454 | 0.05 |
ENSMUST00000115163.3
ENSMUST00000144345.1 ENSMUST00000143682.1 ENSMUST00000115165.3 ENSMUST00000099991.4 ENSMUST00000130410.1 |
Nrros
|
negative regulator of reactive oxygen species |
| chr12_+_79029150 | 0.05 |
ENSMUST00000039928.5
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr8_-_47713920 | 0.05 |
ENSMUST00000038738.5
|
Cdkn2aip
|
CDKN2A interacting protein |
| chr7_-_30626145 | 0.05 |
ENSMUST00000075738.4
|
Cox6b1
|
cytochrome c oxidase, subunit VIb polypeptide 1 |
| chr5_+_121463150 | 0.04 |
ENSMUST00000156080.1
ENSMUST00000031405.5 ENSMUST00000094357.4 |
Tmem116
|
transmembrane protein 116 |
| chr5_-_86518562 | 0.04 |
ENSMUST00000140095.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr10_+_34037597 | 0.04 |
ENSMUST00000095758.1
|
Trappc3l
|
trafficking protein particle complex 3 like |
| chr17_-_8101228 | 0.04 |
ENSMUST00000097422.4
|
Gm1604A
|
predicted gene 1604A |
| chrX_+_160768013 | 0.04 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr9_+_45319072 | 0.04 |
ENSMUST00000034597.7
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr10_-_130112788 | 0.04 |
ENSMUST00000081469.1
|
Olfr823
|
olfactory receptor 823 |
| chr6_-_122340525 | 0.04 |
ENSMUST00000112600.2
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr4_-_141790891 | 0.04 |
ENSMUST00000038014.4
ENSMUST00000153880.1 |
Dnajc16
|
DnaJ (Hsp40) homolog, subfamily C, member 16 |
| chr17_+_34305883 | 0.04 |
ENSMUST00000074557.8
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr18_-_4352944 | 0.04 |
ENSMUST00000025078.2
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr10_+_90829835 | 0.04 |
ENSMUST00000179964.1
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_-_49187037 | 0.03 |
ENSMUST00000153999.1
ENSMUST00000066531.6 |
Btnl9
|
butyrophilin-like 9 |
| chr1_+_158362261 | 0.03 |
ENSMUST00000046110.9
|
Astn1
|
astrotactin 1 |
| chr9_+_109812280 | 0.03 |
ENSMUST00000154663.1
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr10_+_90829780 | 0.03 |
ENSMUST00000179337.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_+_138085083 | 0.03 |
ENSMUST00000019660.4
ENSMUST00000066617.5 ENSMUST00000110963.1 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chrY_+_37100588 | 0.03 |
ENSMUST00000180349.1
|
Gm21799
|
predicted gene, 21799 |
| chr18_-_62756275 | 0.03 |
ENSMUST00000067450.1
ENSMUST00000048109.5 |
2700046A07Rik
|
RIKEN cDNA 2700046A07 gene |
| chr18_-_35740499 | 0.03 |
ENSMUST00000115728.3
|
Tmem173
|
transmembrane protein 173 |
| chr13_+_40859768 | 0.03 |
ENSMUST00000110191.2
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_+_74695601 | 0.03 |
ENSMUST00000152027.1
|
Gm14396
|
predicted gene 14396 |
| chr5_-_125390176 | 0.03 |
ENSMUST00000156249.1
|
Ubc
|
ubiquitin C |
| chr15_+_25742314 | 0.03 |
ENSMUST00000135981.1
|
Myo10
|
myosin X |
| chr10_-_130280218 | 0.03 |
ENSMUST00000061571.3
|
Neurod4
|
neurogenic differentiation 4 |
| chr19_+_26748268 | 0.03 |
ENSMUST00000175791.1
ENSMUST00000176698.1 ENSMUST00000177252.1 ENSMUST00000176475.1 ENSMUST00000112637.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_125318334 | 0.03 |
ENSMUST00000060220.1
|
4930533L02Rik
|
RIKEN cDNA 4930533L02 gene |
| chrX_+_164373363 | 0.03 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr10_-_78753046 | 0.03 |
ENSMUST00000105383.2
|
Ccdc105
|
coiled-coil domain containing 105 |
| chr18_-_38929148 | 0.02 |
ENSMUST00000134864.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr6_-_122340499 | 0.02 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr15_+_79690869 | 0.02 |
ENSMUST00000046463.8
|
Gtpbp1
|
GTP binding protein 1 |
| chr3_+_99141068 | 0.02 |
ENSMUST00000004343.2
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial) |
| chr7_-_103788275 | 0.02 |
ENSMUST00000183254.1
|
Olfr67
|
olfactory receptor 67 |
| chr14_+_52795119 | 0.02 |
ENSMUST00000185019.1
|
Trav6d-5
|
T cell receptor alpha variable 6D-5 |
| chr9_+_107580117 | 0.02 |
ENSMUST00000093785.4
|
Nat6
|
N-acetyltransferase 6 |
| chr7_-_65371210 | 0.02 |
ENSMUST00000102592.3
|
Tjp1
|
tight junction protein 1 |
| chr17_-_33139499 | 0.02 |
ENSMUST00000131954.1
|
Morc2b
|
microrchidia 2B |
| chr9_-_75597643 | 0.02 |
ENSMUST00000164100.1
|
Tmod2
|
tropomodulin 2 |
| chr5_+_66968416 | 0.02 |
ENSMUST00000038188.7
|
Limch1
|
LIM and calponin homology domains 1 |
| chr14_+_73173825 | 0.01 |
ENSMUST00000166875.1
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_+_104095796 | 0.01 |
ENSMUST00000040423.5
ENSMUST00000168176.1 |
Cd59a
|
CD59a antigen |
| chr11_+_105126425 | 0.01 |
ENSMUST00000021030.7
|
Mettl2
|
methyltransferase like 2 |
| chr11_+_106751255 | 0.01 |
ENSMUST00000183111.1
ENSMUST00000106794.2 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr9_-_37712277 | 0.01 |
ENSMUST00000104875.1
|
Olfr160
|
olfactory receptor 160 |
| chr13_+_43370710 | 0.01 |
ENSMUST00000066804.4
|
Sirt5
|
sirtuin 5 |
| chr12_-_56536895 | 0.01 |
ENSMUST00000001536.8
|
Nkx2-1
|
NK2 homeobox 1 |
| chr2_-_66568312 | 0.01 |
ENSMUST00000112354.1
|
Scn9a
|
sodium channel, voltage-gated, type IX, alpha |
| chr7_-_28949670 | 0.00 |
ENSMUST00000148196.1
|
Actn4
|
actinin alpha 4 |
| chr10_+_127421124 | 0.00 |
ENSMUST00000170336.1
|
R3hdm2
|
R3H domain containing 2 |
| chr7_+_19382005 | 0.00 |
ENSMUST00000062831.9
ENSMUST00000108461.1 ENSMUST00000108460.1 |
Ercc2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr5_-_103211251 | 0.00 |
ENSMUST00000060871.5
ENSMUST00000112846.1 ENSMUST00000170792.1 ENSMUST00000112847.2 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr14_-_41008256 | 0.00 |
ENSMUST00000136661.1
|
Fam213a
|
family with sequence similarity 213, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 0.7 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 4.0 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 0.1 | 1.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.4 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) peptide antigen transport(GO:0046968) |
| 0.1 | 0.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 1.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.3 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.3 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.2 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.3 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.0 | 1.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 1.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.7 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.2 | 0.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 0.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.4 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.3 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.1 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 1.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |