Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
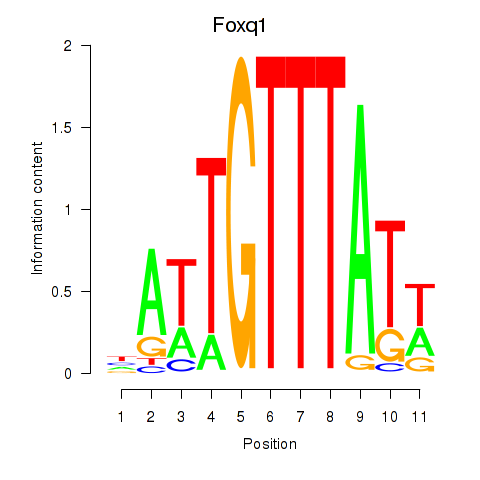
Results for Foxq1
Z-value: 0.89
Transcription factors associated with Foxq1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxq1
|
ENSMUSG00000038415.8 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxq1 | mm10_v2_chr13_+_31558157_31558176 | 0.06 | 7.1e-01 | Click! |
Activity profile of Foxq1 motif
Sorted Z-values of Foxq1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_139563316 | 1.39 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr8_+_45658731 | 1.36 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_+_51117754 | 1.33 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr5_+_146079254 | 1.24 |
ENSMUST00000035571.8
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr8_+_45658666 | 1.22 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_139560158 | 1.18 |
ENSMUST00000160423.1
ENSMUST00000023965.5 |
Cfhr1
|
complement factor H-related 1 |
| chr2_-_77519565 | 1.16 |
ENSMUST00000111830.2
|
Zfp385b
|
zinc finger protein 385B |
| chr5_-_145805862 | 1.15 |
ENSMUST00000067479.5
|
Cyp3a44
|
cytochrome P450, family 3, subfamily a, polypeptide 44 |
| chr6_-_98342728 | 1.12 |
ENSMUST00000164491.1
|
Gm765
|
predicted gene 765 |
| chr13_-_22219820 | 1.11 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr5_-_31108218 | 1.10 |
ENSMUST00000182776.1
ENSMUST00000182444.1 |
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr1_+_51289106 | 1.08 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr5_-_145879857 | 1.06 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr5_-_145469723 | 1.02 |
ENSMUST00000031633.4
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr1_-_139781236 | 0.94 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr3_+_135825648 | 0.88 |
ENSMUST00000180196.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr14_-_36935560 | 0.81 |
ENSMUST00000183038.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr15_+_54410755 | 0.80 |
ENSMUST00000036737.3
|
Colec10
|
collectin sub-family member 10 |
| chr3_+_7366598 | 0.77 |
ENSMUST00000028999.6
|
Pkia
|
protein kinase inhibitor, alpha |
| chr11_-_46389471 | 0.76 |
ENSMUST00000109237.2
|
Itk
|
IL2 inducible T cell kinase |
| chr16_+_43363855 | 0.76 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_-_37703275 | 0.68 |
ENSMUST00000072186.5
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr18_-_39489157 | 0.68 |
ENSMUST00000131885.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr11_-_46389509 | 0.67 |
ENSMUST00000020664.6
|
Itk
|
IL2 inducible T cell kinase |
| chr16_-_88563166 | 0.62 |
ENSMUST00000049697.4
|
Cldn8
|
claudin 8 |
| chr17_+_12584183 | 0.59 |
ENSMUST00000046959.7
|
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr5_+_102481546 | 0.59 |
ENSMUST00000112854.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_+_68584154 | 0.58 |
ENSMUST00000182997.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr12_+_77238093 | 0.55 |
ENSMUST00000177595.1
ENSMUST00000171770.2 |
Fut8
|
fucosyltransferase 8 |
| chr1_-_4360256 | 0.54 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_+_91255954 | 0.53 |
ENSMUST00000134699.1
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_46389454 | 0.53 |
ENSMUST00000101306.3
|
Itk
|
IL2 inducible T cell kinase |
| chr17_+_31433054 | 0.52 |
ENSMUST00000136384.1
|
Pde9a
|
phosphodiesterase 9A |
| chr2_-_91255995 | 0.52 |
ENSMUST00000180732.1
|
Gm17281
|
predicted gene, 17281 |
| chr2_+_91256144 | 0.51 |
ENSMUST00000154959.1
ENSMUST00000059566.4 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr19_+_30232921 | 0.50 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr10_+_34297421 | 0.49 |
ENSMUST00000047935.6
|
Tspyl4
|
TSPY-like 4 |
| chr3_+_19957037 | 0.48 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr7_+_24134148 | 0.48 |
ENSMUST00000056549.7
|
Zfp235
|
zinc finger protein 235 |
| chr11_-_62392605 | 0.48 |
ENSMUST00000151498.2
ENSMUST00000159069.1 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr13_-_21453714 | 0.48 |
ENSMUST00000032820.7
ENSMUST00000110485.1 |
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr4_+_97772734 | 0.46 |
ENSMUST00000152023.1
|
Nfia
|
nuclear factor I/A |
| chr10_+_88091070 | 0.46 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr19_-_17837620 | 0.46 |
ENSMUST00000025618.8
ENSMUST00000050715.8 |
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr2_-_51149100 | 0.46 |
ENSMUST00000154545.1
ENSMUST00000017288.2 |
Rnd3
|
Rho family GTPase 3 |
| chr6_+_97991776 | 0.45 |
ENSMUST00000043628.6
|
Mitf
|
microphthalmia-associated transcription factor |
| chr18_+_69344503 | 0.45 |
ENSMUST00000114985.3
|
Tcf4
|
transcription factor 4 |
| chr19_-_28963863 | 0.45 |
ENSMUST00000161813.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr3_-_146781351 | 0.44 |
ENSMUST00000005164.7
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr10_+_97479470 | 0.44 |
ENSMUST00000105287.3
|
Dcn
|
decorin |
| chr1_+_13668739 | 0.44 |
ENSMUST00000088542.3
|
Xkr9
|
X Kell blood group precursor related family member 9 homolog |
| chrX_+_107149580 | 0.44 |
ENSMUST00000137107.1
ENSMUST00000067249.2 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chr11_-_107348130 | 0.43 |
ENSMUST00000134763.1
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_-_103844173 | 0.43 |
ENSMUST00000022718.3
|
Ednrb
|
endothelin receptor type B |
| chr4_+_34893772 | 0.42 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr4_+_139622842 | 0.42 |
ENSMUST00000039818.9
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr5_+_102481374 | 0.41 |
ENSMUST00000094559.2
ENSMUST00000073302.5 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_+_6184363 | 0.41 |
ENSMUST00000025699.2
ENSMUST00000113528.1 |
1700123I01Rik
|
RIKEN cDNA 1700123I01 gene |
| chr18_+_37477768 | 0.41 |
ENSMUST00000051442.5
|
Pcdhb16
|
protocadherin beta 16 |
| chrX_+_107149454 | 0.40 |
ENSMUST00000125676.1
ENSMUST00000180182.1 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chrX_-_75380041 | 0.39 |
ENSMUST00000114085.2
|
F8
|
coagulation factor VIII |
| chr18_+_11657349 | 0.39 |
ENSMUST00000047322.6
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr4_-_45532470 | 0.39 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr12_+_7977640 | 0.38 |
ENSMUST00000171271.1
ENSMUST00000037811.6 ENSMUST00000037520.7 |
Apob
|
apolipoprotein B |
| chr16_-_85550417 | 0.38 |
ENSMUST00000175700.1
ENSMUST00000114174.2 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr5_+_139543889 | 0.38 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr1_+_43098710 | 0.38 |
ENSMUST00000010434.7
|
AI597479
|
expressed sequence AI597479 |
| chr2_-_148040196 | 0.37 |
ENSMUST00000136555.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr9_-_100506844 | 0.37 |
ENSMUST00000112874.3
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chrX_-_70365052 | 0.37 |
ENSMUST00000101509.2
|
Ids
|
iduronate 2-sulfatase |
| chr5_-_122002340 | 0.35 |
ENSMUST00000134326.1
|
Cux2
|
cut-like homeobox 2 |
| chr19_+_43782181 | 0.35 |
ENSMUST00000026208.4
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr18_+_37489465 | 0.34 |
ENSMUST00000055949.2
|
Pcdhb18
|
protocadherin beta 18 |
| chr18_+_37496997 | 0.33 |
ENSMUST00000059571.5
|
Pcdhb19
|
protocadherin beta 19 |
| chr9_+_15239045 | 0.33 |
ENSMUST00000034413.6
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr11_-_54860564 | 0.32 |
ENSMUST00000144164.1
|
Lyrm7
|
LYR motif containing 7 |
| chr6_-_130233322 | 0.32 |
ENSMUST00000032286.6
|
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr7_-_133782721 | 0.32 |
ENSMUST00000063669.1
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr17_-_32822200 | 0.32 |
ENSMUST00000179695.1
|
Zfp799
|
zinc finger protein 799 |
| chr1_+_33719863 | 0.31 |
ENSMUST00000088287.3
|
Rab23
|
RAB23, member RAS oncogene family |
| chr5_-_146220901 | 0.30 |
ENSMUST00000169407.2
ENSMUST00000161331.1 ENSMUST00000159074.2 ENSMUST00000067837.3 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr15_-_97247287 | 0.30 |
ENSMUST00000053106.5
|
Amigo2
|
adhesion molecule with Ig like domain 2 |
| chr3_+_79884931 | 0.30 |
ENSMUST00000135021.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr9_-_105521147 | 0.30 |
ENSMUST00000176770.1
ENSMUST00000085133.6 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr8_-_84773381 | 0.29 |
ENSMUST00000109764.1
|
Nfix
|
nuclear factor I/X |
| chr3_-_129332713 | 0.29 |
ENSMUST00000029658.7
|
Enpep
|
glutamyl aminopeptidase |
| chr18_+_37447641 | 0.28 |
ENSMUST00000052387.3
|
Pcdhb14
|
protocadherin beta 14 |
| chrM_+_5319 | 0.28 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr13_+_40917626 | 0.28 |
ENSMUST00000067778.6
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr12_+_37242030 | 0.28 |
ENSMUST00000160390.1
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_+_45311538 | 0.28 |
ENSMUST00000087883.6
|
Col3a1
|
collagen, type III, alpha 1 |
| chr14_+_124005355 | 0.26 |
ENSMUST00000166105.1
|
Gm17615
|
predicted gene, 17615 |
| chr1_-_43098622 | 0.26 |
ENSMUST00000095014.1
|
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr12_+_111166485 | 0.26 |
ENSMUST00000139162.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr2_-_126783416 | 0.26 |
ENSMUST00000130356.1
ENSMUST00000028842.2 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr18_-_3281036 | 0.25 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr1_-_170110491 | 0.25 |
ENSMUST00000027985.2
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr3_+_79884496 | 0.25 |
ENSMUST00000118853.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr16_+_43364145 | 0.25 |
ENSMUST00000148775.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_96409038 | 0.24 |
ENSMUST00000179683.1
|
Gm20091
|
predicted gene, 20091 |
| chr5_+_122643878 | 0.24 |
ENSMUST00000100737.3
ENSMUST00000121489.1 ENSMUST00000031425.8 ENSMUST00000086247.5 |
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr7_-_38019505 | 0.24 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr10_+_69925954 | 0.24 |
ENSMUST00000181974.1
ENSMUST00000182795.1 ENSMUST00000182437.1 |
Ank3
|
ankyrin 3, epithelial |
| chr10_+_20148457 | 0.24 |
ENSMUST00000020173.8
|
Map7
|
microtubule-associated protein 7 |
| chr12_-_104865076 | 0.24 |
ENSMUST00000109937.1
ENSMUST00000109936.1 |
Clmn
|
calmin |
| chr19_-_36919606 | 0.24 |
ENSMUST00000057337.7
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr17_+_35077080 | 0.23 |
ENSMUST00000172959.1
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr11_+_77765588 | 0.23 |
ENSMUST00000164315.1
|
Myo18a
|
myosin XVIIIA |
| chr3_+_122419772 | 0.22 |
ENSMUST00000029766.4
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr18_+_37442517 | 0.22 |
ENSMUST00000056915.1
|
Pcdhb13
|
protocadherin beta 13 |
| chr18_-_88927447 | 0.21 |
ENSMUST00000147313.1
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr1_+_156838915 | 0.21 |
ENSMUST00000111720.1
|
Angptl1
|
angiopoietin-like 1 |
| chr11_+_97029925 | 0.21 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr11_+_97030130 | 0.20 |
ENSMUST00000153482.1
|
Scrn2
|
secernin 2 |
| chr19_+_45149833 | 0.19 |
ENSMUST00000026236.9
|
Tlx1
|
T cell leukemia, homeobox 1 |
| chr4_+_109343029 | 0.19 |
ENSMUST00000030281.5
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr14_+_53763083 | 0.19 |
ENSMUST00000180859.1
ENSMUST00000103589.4 |
Trav14-3
|
T cell receptor alpha variable 14-3 |
| chr4_+_108479081 | 0.18 |
ENSMUST00000155068.1
|
Zcchc11
|
zinc finger, CCHC domain containing 11 |
| chrX_-_157568983 | 0.18 |
ENSMUST00000065806.4
|
Yy2
|
Yy2 transcription factor |
| chr9_-_36797303 | 0.18 |
ENSMUST00000115086.5
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr1_+_87404916 | 0.18 |
ENSMUST00000173152.1
ENSMUST00000173663.1 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr5_-_73630303 | 0.17 |
ENSMUST00000152215.1
|
Lrrc66
|
leucine rich repeat containing 66 |
| chrX_-_103483205 | 0.17 |
ENSMUST00000127786.2
|
Xist
|
inactive X specific transcripts |
| chr3_+_96645579 | 0.16 |
ENSMUST00000119365.1
ENSMUST00000029744.5 |
Itga10
|
integrin, alpha 10 |
| chr3_-_36053512 | 0.16 |
ENSMUST00000166644.2
ENSMUST00000062056.8 |
Ccdc144b
|
coiled-coil domain containing 144B |
| chr17_+_35076902 | 0.16 |
ENSMUST00000172494.1
ENSMUST00000172678.1 ENSMUST00000013910.4 |
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr12_+_111166536 | 0.16 |
ENSMUST00000060274.6
|
Traf3
|
TNF receptor-associated factor 3 |
| chr5_-_145720124 | 0.16 |
ENSMUST00000094111.4
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr3_+_145987835 | 0.16 |
ENSMUST00000039517.6
|
Syde2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
| chr16_-_56024628 | 0.16 |
ENSMUST00000119981.1
ENSMUST00000096021.3 |
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr11_+_82911253 | 0.15 |
ENSMUST00000164945.1
ENSMUST00000018989.7 |
Unc45b
|
unc-45 homolog B (C. elegans) |
| chr16_+_13358375 | 0.14 |
ENSMUST00000149359.1
|
Mkl2
|
MKL/myocardin-like 2 |
| chr16_-_26526744 | 0.14 |
ENSMUST00000165687.1
|
Tmem207
|
transmembrane protein 207 |
| chr14_+_52753367 | 0.14 |
ENSMUST00000180717.1
ENSMUST00000183820.1 |
Trav6d-4
|
T cell receptor alpha variable 6D-4 |
| chr8_-_3854309 | 0.13 |
ENSMUST00000033888.4
|
Cd209e
|
CD209e antigen |
| chr7_+_24270420 | 0.13 |
ENSMUST00000108438.3
|
Zfp93
|
zinc finger protein 93 |
| chr4_-_15945359 | 0.13 |
ENSMUST00000029877.8
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr11_-_100822525 | 0.12 |
ENSMUST00000107358.2
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr12_+_110841021 | 0.12 |
ENSMUST00000181348.1
|
4921507G05Rik
|
RIKEN cDNA 4921507G05 gene |
| chr3_+_99885383 | 0.12 |
ENSMUST00000164539.1
|
Spag17
|
sperm associated antigen 17 |
| chr13_-_67637776 | 0.12 |
ENSMUST00000012314.8
|
A530054K11Rik
|
RIKEN cDNA A530054K11 gene |
| chr17_+_20570362 | 0.11 |
ENSMUST00000095633.3
|
Gm5145
|
predicted pseudogene 5145 |
| chr12_-_101958148 | 0.10 |
ENSMUST00000159883.1
ENSMUST00000160251.1 ENSMUST00000161011.1 ENSMUST00000021606.5 |
Atxn3
|
ataxin 3 |
| chr1_-_44101661 | 0.10 |
ENSMUST00000152239.1
|
Tex30
|
testis expressed 30 |
| chr10_-_107486077 | 0.09 |
ENSMUST00000000445.1
|
Myf5
|
myogenic factor 5 |
| chr1_-_195131536 | 0.09 |
ENSMUST00000075451.6
|
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr8_+_78509319 | 0.09 |
ENSMUST00000034111.8
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr6_-_129237948 | 0.09 |
ENSMUST00000181238.1
ENSMUST00000180379.1 |
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene |
| chr2_+_60209887 | 0.09 |
ENSMUST00000102748.4
ENSMUST00000102747.1 |
March7
|
membrane-associated ring finger (C3HC4) 7 |
| chr3_+_136670076 | 0.09 |
ENSMUST00000070198.7
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr2_-_176917518 | 0.09 |
ENSMUST00000108931.2
|
Gm14296
|
predicted gene 14296 |
| chr2_+_112284561 | 0.08 |
ENSMUST00000053666.7
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr11_-_99620432 | 0.08 |
ENSMUST00000073853.2
|
Gm11562
|
predicted gene 11562 |
| chr6_+_117907795 | 0.07 |
ENSMUST00000167657.1
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chrX_-_142196917 | 0.07 |
ENSMUST00000042530.3
|
Gucy2f
|
guanylate cyclase 2f |
| chr9_+_45319072 | 0.07 |
ENSMUST00000034597.7
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr2_+_152669461 | 0.07 |
ENSMUST00000125366.1
ENSMUST00000109825.1 ENSMUST00000089059.2 ENSMUST00000079247.3 |
H13
|
histocompatibility 13 |
| chr4_+_32243733 | 0.07 |
ENSMUST00000165661.1
|
D130062J21Rik
|
RIKEN cDNA D130062J21 gene |
| chr14_-_30353468 | 0.07 |
ENSMUST00000112249.1
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr8_-_67515606 | 0.07 |
ENSMUST00000032981.5
|
Gm9755
|
predicted pseudogene 9755 |
| chr8_-_53638945 | 0.06 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chr13_+_96082158 | 0.06 |
ENSMUST00000185178.1
|
Gm17190
|
predicted gene 17190 |
| chr10_+_96616998 | 0.06 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr13_-_67061131 | 0.06 |
ENSMUST00000167565.1
|
Zfp712
|
zinc finger protein 712 |
| chr5_-_130255525 | 0.06 |
ENSMUST00000026387.4
|
Sbds
|
Shwachman-Bodian-Diamond syndrome homolog (human) |
| chr10_+_42860543 | 0.06 |
ENSMUST00000157071.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr7_+_141476374 | 0.05 |
ENSMUST00000117634.1
|
Tspan4
|
tetraspanin 4 |
| chr1_-_13372434 | 0.05 |
ENSMUST00000081713.4
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr17_-_34118476 | 0.05 |
ENSMUST00000095347.5
|
Brd2
|
bromodomain containing 2 |
| chr5_+_107597760 | 0.05 |
ENSMUST00000112655.1
|
Rpap2
|
RNA polymerase II associated protein 2 |
| chr2_-_6722187 | 0.04 |
ENSMUST00000182657.1
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr5_+_21737141 | 0.04 |
ENSMUST00000030882.5
|
Pmpcb
|
peptidase (mitochondrial processing) beta |
| chrX_-_9469288 | 0.04 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr7_+_120677579 | 0.04 |
ENSMUST00000060175.6
|
BC030336
|
cDNA sequence BC030336 |
| chr11_-_99493112 | 0.04 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr2_-_37647199 | 0.04 |
ENSMUST00000028279.3
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr3_-_57301919 | 0.03 |
ENSMUST00000029376.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr4_+_107314363 | 0.03 |
ENSMUST00000075693.5
ENSMUST00000139527.1 |
Yipf1
|
Yip1 domain family, member 1 |
| chr10_+_24049482 | 0.03 |
ENSMUST00000071691.2
|
Taar7f
|
trace amine-associated receptor 7F |
| chr17_-_40319205 | 0.03 |
ENSMUST00000026498.4
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr5_+_107597696 | 0.03 |
ENSMUST00000112651.1
ENSMUST00000112654.1 ENSMUST00000065422.5 |
Rpap2
|
RNA polymerase II associated protein 2 |
| chr4_+_107314399 | 0.03 |
ENSMUST00000128284.1
ENSMUST00000124650.1 |
Yipf1
|
Yip1 domain family, member 1 |
| chr10_-_130112788 | 0.02 |
ENSMUST00000081469.1
|
Olfr823
|
olfactory receptor 823 |
| chr5_+_135725713 | 0.02 |
ENSMUST00000127096.1
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr5_+_104202609 | 0.01 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr5_-_99252839 | 0.01 |
ENSMUST00000168092.1
ENSMUST00000031276.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr5_-_76331102 | 0.01 |
ENSMUST00000122213.1
|
Pdcl2
|
phosducin-like 2 |
| chr2_+_35224516 | 0.01 |
ENSMUST00000124489.1
|
Gm13605
|
predicted gene 13605 |
| chr19_-_56822161 | 0.01 |
ENSMUST00000118592.1
|
A630007B06Rik
|
RIKEN cDNA A630007B06 gene |
| chr10_+_60106452 | 0.01 |
ENSMUST00000165024.2
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr6_-_48086530 | 0.01 |
ENSMUST00000073124.6
|
Zfp746
|
zinc finger protein 746 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxq1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | GO:0050787 | detoxification of mercury ion(GO:0050787) detoxification of inorganic compound(GO:0061687) |
| 0.2 | 0.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.5 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 1.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 2.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.7 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 1.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.1 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.9 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) regulation of bleb assembly(GO:1904170) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.3 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.2 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 2.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.4 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 1.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.4 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.7 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0098576 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) lumenal side of membrane(GO:0098576) |
| 0.0 | 2.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.6 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.2 | 0.6 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.7 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.3 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 1.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |