Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
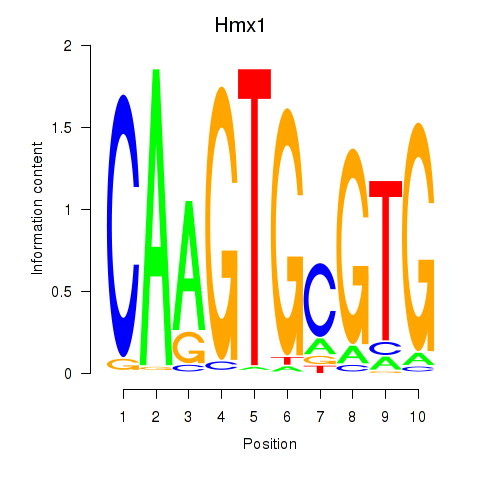
Results for Hmx1
Z-value: 2.25
Transcription factors associated with Hmx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx1
|
ENSMUSG00000067438.3 | H6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx1 | mm10_v2_chr5_+_35388844_35388882 | 0.32 | 6.0e-02 | Click! |
Activity profile of Hmx1 motif
Sorted Z-values of Hmx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_56102458 | 16.48 |
ENSMUST00000015583.1
|
Ctsg
|
cathepsin G |
| chr12_+_109459843 | 12.74 |
ENSMUST00000173812.1
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr14_-_79301623 | 10.44 |
ENSMUST00000022595.7
|
Rgcc
|
regulator of cell cycle |
| chr7_+_18884679 | 8.14 |
ENSMUST00000032573.6
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr4_+_120666562 | 6.98 |
ENSMUST00000094814.4
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr17_-_33890539 | 6.74 |
ENSMUST00000173386.1
|
Kifc1
|
kinesin family member C1 |
| chr6_+_30738044 | 6.68 |
ENSMUST00000128398.1
ENSMUST00000163949.2 ENSMUST00000124665.1 |
Mest
|
mesoderm specific transcript |
| chr13_+_91461050 | 6.61 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr17_-_33890584 | 6.46 |
ENSMUST00000114361.2
ENSMUST00000173492.1 |
Kifc1
|
kinesin family member C1 |
| chr11_+_69965396 | 6.27 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr8_-_122460666 | 5.65 |
ENSMUST00000006762.5
|
Snai3
|
snail homolog 3 (Drosophila) |
| chr1_-_75505641 | 5.54 |
ENSMUST00000155084.1
|
Obsl1
|
obscurin-like 1 |
| chr1_-_75506331 | 5.24 |
ENSMUST00000113567.2
ENSMUST00000113565.2 |
Obsl1
|
obscurin-like 1 |
| chr11_+_61956779 | 5.10 |
ENSMUST00000049836.7
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chrX_+_50841434 | 5.08 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chr6_+_90619241 | 5.08 |
ENSMUST00000032177.8
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr4_-_152477433 | 5.02 |
ENSMUST00000159186.1
ENSMUST00000162017.1 ENSMUST00000030768.2 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr17_-_71310952 | 4.94 |
ENSMUST00000024849.9
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr1_+_43730593 | 4.83 |
ENSMUST00000027217.8
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene |
| chr11_+_11686213 | 4.79 |
ENSMUST00000076700.4
ENSMUST00000048122.6 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr6_+_29433248 | 4.78 |
ENSMUST00000101617.2
ENSMUST00000065090.5 |
Flnc
|
filamin C, gamma |
| chr17_+_26917091 | 4.76 |
ENSMUST00000078961.4
|
Kifc5b
|
kinesin family member C5B |
| chr14_-_56234650 | 4.51 |
ENSMUST00000015585.2
|
Gzmc
|
granzyme C |
| chr3_-_152166230 | 4.51 |
ENSMUST00000046614.9
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chrX_-_7671341 | 4.42 |
ENSMUST00000033486.5
|
Plp2
|
proteolipid protein 2 |
| chr6_-_67535783 | 4.41 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr2_+_153492790 | 4.20 |
ENSMUST00000109783.1
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr12_+_35992900 | 4.18 |
ENSMUST00000020898.5
|
Agr2
|
anterior gradient 2 |
| chr6_+_29433131 | 3.81 |
ENSMUST00000090474.4
|
Flnc
|
filamin C, gamma |
| chr3_+_28781305 | 3.75 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chrX_-_134808984 | 3.74 |
ENSMUST00000035559.4
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr9_-_42124276 | 3.67 |
ENSMUST00000060989.8
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr9_+_108560422 | 3.64 |
ENSMUST00000081111.8
|
Impdh2
|
inosine 5'-phosphate dehydrogenase 2 |
| chr2_-_92370999 | 3.56 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr14_+_54640952 | 3.52 |
ENSMUST00000169818.2
|
Gm17606
|
predicted gene, 17606 |
| chr12_+_110279228 | 3.48 |
ENSMUST00000097228.4
|
Dio3
|
deiodinase, iodothyronine type III |
| chr2_-_92371039 | 3.47 |
ENSMUST00000068586.6
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr11_-_46312220 | 3.41 |
ENSMUST00000129474.1
ENSMUST00000093166.4 ENSMUST00000165599.2 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr11_+_3290300 | 3.34 |
ENSMUST00000057089.6
ENSMUST00000093402.5 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chrX_-_74246364 | 3.26 |
ENSMUST00000130007.1
|
Flna
|
filamin, alpha |
| chr2_+_129198757 | 3.23 |
ENSMUST00000028880.3
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr8_+_105297663 | 3.22 |
ENSMUST00000015003.8
|
E2f4
|
E2F transcription factor 4 |
| chrX_-_74246534 | 3.22 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr7_-_126369543 | 3.17 |
ENSMUST00000032997.6
|
Lat
|
linker for activation of T cells |
| chr10_-_61273242 | 3.12 |
ENSMUST00000120336.1
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr6_-_124733067 | 3.04 |
ENSMUST00000173647.1
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_-_127151044 | 3.04 |
ENSMUST00000000188.8
|
Ccnd2
|
cyclin D2 |
| chr2_+_92915080 | 3.01 |
ENSMUST00000028648.2
|
Syt13
|
synaptotagmin XIII |
| chr2_-_92370968 | 2.99 |
ENSMUST00000176774.1
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr17_-_23684019 | 2.92 |
ENSMUST00000085989.5
|
Cldn9
|
claudin 9 |
| chr3_+_145292472 | 2.87 |
ENSMUST00000029848.4
ENSMUST00000139001.1 |
Col24a1
|
collagen, type XXIV, alpha 1 |
| chr13_+_55369732 | 2.87 |
ENSMUST00000063771.7
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr15_-_79834261 | 2.80 |
ENSMUST00000148358.1
|
Cbx6
|
chromobox 6 |
| chr15_-_78572754 | 2.77 |
ENSMUST00000043214.6
|
Rac2
|
RAS-related C3 botulinum substrate 2 |
| chr15_-_79834224 | 2.72 |
ENSMUST00000109623.1
ENSMUST00000109625.1 ENSMUST00000023060.6 ENSMUST00000089299.5 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr6_+_34476207 | 2.70 |
ENSMUST00000045372.5
ENSMUST00000138668.1 ENSMUST00000139067.1 |
Bpgm
|
2,3-bisphosphoglycerate mutase |
| chr1_+_52008210 | 2.69 |
ENSMUST00000027277.5
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr18_-_62179948 | 2.68 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr17_+_48462355 | 2.68 |
ENSMUST00000162132.1
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like |
| chr18_+_62180119 | 2.67 |
ENSMUST00000067743.1
|
Gm9949
|
predicted gene 9949 |
| chr7_+_118712516 | 2.63 |
ENSMUST00000106557.1
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr5_+_121220191 | 2.61 |
ENSMUST00000119892.2
ENSMUST00000042614.6 |
Gm15800
|
predicted gene 15800 |
| chr2_-_73453918 | 2.56 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_-_127499095 | 2.54 |
ENSMUST00000182594.1
|
Ank2
|
ankyrin 2, brain |
| chr4_+_129985098 | 2.43 |
ENSMUST00000106017.1
ENSMUST00000121049.1 |
Bai2
|
brain-specific angiogenesis inhibitor 2 |
| chr9_-_61976563 | 2.43 |
ENSMUST00000113990.1
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr19_-_4615453 | 2.37 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_-_69858723 | 2.34 |
ENSMUST00000001626.3
ENSMUST00000108626.1 |
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr9_+_62838767 | 2.32 |
ENSMUST00000034776.6
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr8_+_88272403 | 2.28 |
ENSMUST00000169037.1
|
Adcy7
|
adenylate cyclase 7 |
| chr4_+_94556546 | 2.25 |
ENSMUST00000094969.1
|
Gm10306
|
predicted gene 10306 |
| chr8_+_70315759 | 2.24 |
ENSMUST00000165819.2
ENSMUST00000140239.1 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chr10_-_61273409 | 2.24 |
ENSMUST00000092486.4
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr16_+_49699198 | 2.23 |
ENSMUST00000046777.4
ENSMUST00000142682.1 |
Ift57
|
intraflagellar transport 57 |
| chr17_-_47833169 | 2.20 |
ENSMUST00000131971.1
ENSMUST00000129360.1 ENSMUST00000113280.1 ENSMUST00000132125.1 |
Mdfi
|
MyoD family inhibitor |
| chr11_+_44518959 | 2.19 |
ENSMUST00000019333.3
|
Rnf145
|
ring finger protein 145 |
| chr11_+_68692070 | 2.18 |
ENSMUST00000108673.1
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr17_-_47833256 | 2.16 |
ENSMUST00000152455.1
ENSMUST00000035375.7 |
Mdfi
|
MyoD family inhibitor |
| chr16_+_11984581 | 2.12 |
ENSMUST00000170672.2
ENSMUST00000023138.7 |
Shisa9
|
shisa homolog 9 (Xenopus laevis) |
| chr3_-_92485886 | 2.11 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr2_+_165595009 | 2.11 |
ENSMUST00000088132.6
|
Eya2
|
eyes absent 2 homolog (Drosophila) |
| chr10_-_76961788 | 2.10 |
ENSMUST00000001148.4
ENSMUST00000105411.2 |
Pcbp3
|
poly(rC) binding protein 3 |
| chr3_-_89418287 | 2.10 |
ENSMUST00000029679.3
|
Cks1b
|
CDC28 protein kinase 1b |
| chr13_+_20090538 | 2.09 |
ENSMUST00000072519.5
|
Elmo1
|
engulfment and cell motility 1 |
| chr10_+_80264942 | 2.08 |
ENSMUST00000105362.1
ENSMUST00000105361.3 |
Dazap1
|
DAZ associated protein 1 |
| chr8_+_120537423 | 2.08 |
ENSMUST00000118136.1
|
Gse1
|
genetic suppressor element 1 |
| chrX_+_159255919 | 2.02 |
ENSMUST00000112492.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr9_-_21312255 | 1.97 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr7_-_79715669 | 1.97 |
ENSMUST00000184137.1
ENSMUST00000183846.1 |
Kif7
|
kinesin family member 7 |
| chr3_+_108085976 | 1.96 |
ENSMUST00000070502.1
|
Gm12500
|
predicted gene 12500 |
| chr15_+_54745702 | 1.96 |
ENSMUST00000050027.8
|
Nov
|
nephroblastoma overexpressed gene |
| chr2_+_130274437 | 1.95 |
ENSMUST00000141872.1
|
Nop56
|
NOP56 ribonucleoprotein |
| chrY_+_90843934 | 1.91 |
ENSMUST00000178550.1
|
Gm21742
|
predicted gene, 21742 |
| chr11_+_3289880 | 1.90 |
ENSMUST00000110043.1
ENSMUST00000094471.3 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr15_-_98778150 | 1.89 |
ENSMUST00000023732.5
|
Wnt10b
|
wingless related MMTV integration site 10b |
| chr5_-_140565060 | 1.89 |
ENSMUST00000042993.6
|
Grifin
|
galectin-related inter-fiber protein |
| chr11_+_68691906 | 1.88 |
ENSMUST00000102611.3
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr2_+_153108468 | 1.83 |
ENSMUST00000109799.1
ENSMUST00000003370.7 |
Hck
|
hemopoietic cell kinase |
| chr18_-_24603791 | 1.82 |
ENSMUST00000070726.3
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr13_+_20090500 | 1.80 |
ENSMUST00000165249.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr8_+_94152607 | 1.79 |
ENSMUST00000034211.8
|
Mt3
|
metallothionein 3 |
| chr1_+_135729147 | 1.79 |
ENSMUST00000027677.7
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr2_+_25242227 | 1.77 |
ENSMUST00000154498.1
|
Rnf208
|
ring finger protein 208 |
| chr11_+_77982710 | 1.76 |
ENSMUST00000108360.1
ENSMUST00000049167.7 |
Phf12
|
PHD finger protein 12 |
| chr13_-_32338565 | 1.76 |
ENSMUST00000041859.7
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr7_-_83735503 | 1.73 |
ENSMUST00000001792.4
|
Il16
|
interleukin 16 |
| chr7_-_126160992 | 1.73 |
ENSMUST00000164741.1
|
Xpo6
|
exportin 6 |
| chr18_+_65581704 | 1.71 |
ENSMUST00000182979.1
|
Zfp532
|
zinc finger protein 532 |
| chr17_+_34592248 | 1.71 |
ENSMUST00000038149.6
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr5_-_134614953 | 1.70 |
ENSMUST00000036362.6
ENSMUST00000077636.4 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr15_-_102231920 | 1.69 |
ENSMUST00000001327.3
ENSMUST00000127014.1 |
Itgb7
|
integrin beta 7 |
| chr19_-_4615647 | 1.68 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chrX_-_166585679 | 1.68 |
ENSMUST00000000412.2
|
Egfl6
|
EGF-like-domain, multiple 6 |
| chr15_+_102503722 | 1.67 |
ENSMUST00000096145.4
|
Gm10337
|
predicted gene 10337 |
| chr15_-_74709535 | 1.65 |
ENSMUST00000050234.2
|
Jrk
|
jerky |
| chr6_-_137169678 | 1.64 |
ENSMUST00000119610.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr2_+_32876114 | 1.64 |
ENSMUST00000028135.8
|
Fam129b
|
family with sequence similarity 129, member B |
| chr1_-_87156127 | 1.59 |
ENSMUST00000160810.1
|
Ecel1
|
endothelin converting enzyme-like 1 |
| chr3_+_131112785 | 1.55 |
ENSMUST00000098611.3
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr4_+_129984833 | 1.53 |
ENSMUST00000120204.1
|
Bai2
|
brain-specific angiogenesis inhibitor 2 |
| chr14_-_54686926 | 1.53 |
ENSMUST00000022793.8
ENSMUST00000111484.2 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr11_+_68692097 | 1.50 |
ENSMUST00000018887.8
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr6_-_91116785 | 1.48 |
ENSMUST00000113509.1
ENSMUST00000032179.7 |
Nup210
|
nucleoporin 210 |
| chr2_-_38287174 | 1.47 |
ENSMUST00000130472.1
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr4_+_134468320 | 1.47 |
ENSMUST00000030636.4
ENSMUST00000127279.1 ENSMUST00000105867.1 |
Stmn1
|
stathmin 1 |
| chr8_+_83389846 | 1.44 |
ENSMUST00000002259.6
|
Clgn
|
calmegin |
| chr6_+_86849488 | 1.43 |
ENSMUST00000089519.6
ENSMUST00000113668.1 |
Aak1
|
AP2 associated kinase 1 |
| chr3_-_56183678 | 1.41 |
ENSMUST00000029374.6
|
Nbea
|
neurobeachin |
| chr2_+_130274424 | 1.40 |
ENSMUST00000103198.4
|
Nop56
|
NOP56 ribonucleoprotein |
| chr4_+_140986873 | 1.38 |
ENSMUST00000168047.1
ENSMUST00000037055.7 ENSMUST00000127833.2 |
Atp13a2
|
ATPase type 13A2 |
| chr8_+_83389878 | 1.37 |
ENSMUST00000109831.2
|
Clgn
|
calmegin |
| chrX_+_9272756 | 1.36 |
ENSMUST00000015486.6
|
Xk
|
Kell blood group precursor (McLeod phenotype) homolog |
| chr17_-_36867187 | 1.36 |
ENSMUST00000025329.6
ENSMUST00000174195.1 |
Trim15
|
tripartite motif-containing 15 |
| chr15_+_32244801 | 1.36 |
ENSMUST00000067458.6
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr11_+_115974709 | 1.31 |
ENSMUST00000021107.7
ENSMUST00000106461.1 ENSMUST00000169928.1 |
Itgb4
|
integrin beta 4 |
| chr9_+_32696005 | 1.29 |
ENSMUST00000034534.6
ENSMUST00000050797.7 ENSMUST00000184887.1 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr15_-_79141197 | 1.29 |
ENSMUST00000169604.1
|
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr14_-_54686605 | 1.28 |
ENSMUST00000147714.1
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr11_-_69858687 | 1.28 |
ENSMUST00000125571.1
|
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr15_+_103503261 | 1.28 |
ENSMUST00000023132.3
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr1_+_166130467 | 1.24 |
ENSMUST00000166860.1
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr15_-_79834323 | 1.23 |
ENSMUST00000177316.2
ENSMUST00000175858.2 |
Nptxr
|
neuronal pentraxin receptor |
| chr11_+_62820469 | 1.23 |
ENSMUST00000108703.1
|
Trim16
|
tripartite motif-containing 16 |
| chr10_+_80265035 | 1.22 |
ENSMUST00000092305.5
|
Dazap1
|
DAZ associated protein 1 |
| chr8_+_83900706 | 1.18 |
ENSMUST00000045393.8
ENSMUST00000132500.1 ENSMUST00000152978.1 |
Lphn1
|
latrophilin 1 |
| chr1_+_166130238 | 1.18 |
ENSMUST00000060833.7
ENSMUST00000166159.1 |
Gpa33
|
glycoprotein A33 (transmembrane) |
| chrX_+_159255782 | 1.16 |
ENSMUST00000126686.1
ENSMUST00000033671.6 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr5_-_39644597 | 1.12 |
ENSMUST00000152057.1
|
Hs3st1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr7_-_46179929 | 1.09 |
ENSMUST00000033123.6
|
Abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr2_-_162661075 | 1.08 |
ENSMUST00000109442.1
ENSMUST00000109445.2 ENSMUST00000109443.1 ENSMUST00000109441.1 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr9_+_106170918 | 1.07 |
ENSMUST00000020490.5
|
Wdr82
|
WD repeat domain containing 82 |
| chr15_-_32244632 | 1.05 |
ENSMUST00000181536.1
|
0610007N19Rik
|
RIKEN cDNA 0610007N19 |
| chr11_+_68432112 | 1.05 |
ENSMUST00000021283.7
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5, p101 |
| chr11_-_94242701 | 1.04 |
ENSMUST00000061469.3
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr15_-_13173607 | 1.04 |
ENSMUST00000036439.4
|
Cdh6
|
cadherin 6 |
| chr18_+_34220890 | 1.03 |
ENSMUST00000171187.1
|
Apc
|
adenomatosis polyposis coli |
| chr17_-_24073479 | 1.03 |
ENSMUST00000017090.5
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr2_-_164404606 | 1.02 |
ENSMUST00000109359.1
ENSMUST00000109358.1 ENSMUST00000103103.3 |
Matn4
|
matrilin 4 |
| chr15_+_59374198 | 1.01 |
ENSMUST00000079703.3
ENSMUST00000168722.1 |
Nsmce2
|
non-SMC element 2 homolog (MMS21, S. cerevisiae) |
| chr6_-_137169710 | 1.00 |
ENSMUST00000117919.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr1_-_153186447 | 0.98 |
ENSMUST00000027753.6
|
Lamc2
|
laminin, gamma 2 |
| chr14_-_52237791 | 0.97 |
ENSMUST00000149975.1
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr5_+_100518309 | 0.96 |
ENSMUST00000045993.8
ENSMUST00000151414.1 |
Cops4
|
COP9 (constitutive photomorphogenic) homolog, subunit 4 (Arabidopsis thaliana) |
| chr15_-_79687776 | 0.96 |
ENSMUST00000023061.5
|
Josd1
|
Josephin domain containing 1 |
| chr5_-_33274966 | 0.96 |
ENSMUST00000079746.6
|
Ctbp1
|
C-terminal binding protein 1 |
| chr2_+_118926496 | 0.92 |
ENSMUST00000099546.4
ENSMUST00000110837.1 |
Chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr15_-_85578070 | 0.91 |
ENSMUST00000109424.2
|
Wnt7b
|
wingless-related MMTV integration site 7B |
| chr9_-_44735189 | 0.91 |
ENSMUST00000034611.8
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr2_+_28513105 | 0.90 |
ENSMUST00000135803.1
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr11_+_115974930 | 0.90 |
ENSMUST00000106460.2
|
Itgb4
|
integrin beta 4 |
| chr11_-_102319093 | 0.89 |
ENSMUST00000174302.1
ENSMUST00000178839.1 ENSMUST00000006754.7 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr5_-_139130159 | 0.88 |
ENSMUST00000129851.1
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr17_+_86963279 | 0.87 |
ENSMUST00000139344.1
|
Rhoq
|
ras homolog gene family, member Q |
| chr4_+_116221689 | 0.86 |
ENSMUST00000106490.2
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr5_+_147430407 | 0.84 |
ENSMUST00000176600.1
|
Pan3
|
PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae) |
| chr9_-_110654161 | 0.84 |
ENSMUST00000133191.1
ENSMUST00000167320.1 |
Nbeal2
|
neurobeachin-like 2 |
| chr17_-_12769605 | 0.81 |
ENSMUST00000024599.7
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr5_+_115429944 | 0.79 |
ENSMUST00000067168.5
|
Msi1
|
musashi RNA-binding protein 1 |
| chr13_-_38658991 | 0.78 |
ENSMUST00000001757.7
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr15_+_60822947 | 0.78 |
ENSMUST00000180730.1
|
9930014A18Rik
|
RIKEN cDNA 9930014A18 gene |
| chrX_-_74373260 | 0.77 |
ENSMUST00000073067.4
ENSMUST00000037967.5 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr1_-_143702832 | 0.77 |
ENSMUST00000018337.7
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chr3_-_108085346 | 0.77 |
ENSMUST00000078912.5
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr5_-_39644634 | 0.76 |
ENSMUST00000053116.6
|
Hs3st1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr11_-_116110211 | 0.76 |
ENSMUST00000106441.1
ENSMUST00000021120.5 |
Trim47
|
tripartite motif-containing 47 |
| chr17_-_35979679 | 0.76 |
ENSMUST00000173724.1
ENSMUST00000172900.1 ENSMUST00000174849.1 |
Prr3
|
proline-rich polypeptide 3 |
| chr10_-_80844025 | 0.75 |
ENSMUST00000053986.7
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr12_-_24493656 | 0.73 |
ENSMUST00000073088.2
|
Gm16372
|
predicted pseudogene 16372 |
| chr2_+_28192971 | 0.72 |
ENSMUST00000113920.1
|
Olfm1
|
olfactomedin 1 |
| chr4_+_156013835 | 0.70 |
ENSMUST00000030952.5
|
Tnfrsf4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr8_+_82863351 | 0.69 |
ENSMUST00000078525.5
|
Rnf150
|
ring finger protein 150 |
| chr2_-_119271202 | 0.69 |
ENSMUST00000037360.7
|
Rhov
|
ras homolog gene family, member V |
| chr14_-_52237572 | 0.69 |
ENSMUST00000089752.4
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr17_-_33824346 | 0.67 |
ENSMUST00000173879.1
ENSMUST00000166693.2 ENSMUST00000173019.1 ENSMUST00000087342.6 ENSMUST00000173844.1 |
Rps28
|
ribosomal protein S28 |
| chr5_-_34288318 | 0.66 |
ENSMUST00000094868.3
|
Zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr15_-_79254737 | 0.65 |
ENSMUST00000039752.3
|
Slc16a8
|
solute carrier family 16 (monocarboxylic acid transporters), member 8 |
| chr7_-_30914327 | 0.63 |
ENSMUST00000040548.7
|
Mag
|
myelin-associated glycoprotein |
| chr17_+_35194502 | 0.61 |
ENSMUST00000173600.1
|
Ltb
|
lymphotoxin B |
| chr1_-_9700209 | 0.61 |
ENSMUST00000088658.4
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr3_-_88548249 | 0.61 |
ENSMUST00000131775.1
ENSMUST00000008745.6 |
Rab25
|
RAB25, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 3.5 | 10.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.7 | 8.1 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 2.2 | 6.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.9 | 5.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.8 | 16.5 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 1.6 | 4.8 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.0 | 4.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.9 | 3.7 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.9 | 4.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.9 | 1.8 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.7 | 2.2 | GO:0071492 | cellular response to UV-A(GO:0071492) response to dithiothreitol(GO:0072720) |
| 0.7 | 3.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.7 | 3.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.7 | 10.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 2.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.7 | 2.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.7 | 2.0 | GO:0071603 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 0.6 | 3.8 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 2.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 4.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.5 | 2.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.5 | 1.9 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.4 | 3.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 12.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.4 | 1.6 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.4 | 5.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.4 | 1.1 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.4 | 2.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 3.5 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.3 | 1.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.3 | 1.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 | 6.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 1.0 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 0.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 0.9 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.3 | 2.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.4 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 2.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 2.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.3 | 2.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.3 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.2 | 1.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 3.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 3.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.4 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 1.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 0.8 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 2.2 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 1.8 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 4.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 1.3 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 | 0.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 2.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 7.0 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 3.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 8.3 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 5.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 3.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 2.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 1.6 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 3.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 5.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 2.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 2.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 3.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.6 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 0.8 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.6 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 1.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.9 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 1.6 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 1.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 3.9 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 1.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.9 | GO:1903077 | cortical actin cytoskeleton organization(GO:0030866) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 3.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.1 | GO:1905034 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 2.4 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.0 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 4.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 2.6 | GO:0030217 | T cell differentiation(GO:0030217) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.9 | 5.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 3.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.6 | 12.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 3.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 10.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 3.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 1.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.3 | 2.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 4.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 4.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 1.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 4.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 5.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 3.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 2.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 14.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 1.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 3.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 7.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 5.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 5.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.0 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 3.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 4.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.8 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 3.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 5.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 8.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 18.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 1.3 | 6.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.2 | 3.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.1 | 3.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.9 | 3.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.9 | 2.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.7 | 2.7 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.6 | 10.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 2.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.5 | 3.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.5 | 13.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 1.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 2.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 2.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.3 | 3.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 2.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.3 | 5.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 4.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 10.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 8.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.6 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 3.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 5.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 4.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 0.9 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.2 | 1.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 5.6 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 3.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 2.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 2.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 2.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 7.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.3 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 7.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 18.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 5.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 2.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex RNA binding(GO:0002151) G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 5.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 6.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 3.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 3.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 5.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 15.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 3.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 3.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 13.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 8.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 2.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 1.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 4.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 4.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 13.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 12.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 8.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 7.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 7.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 3.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 3.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 5.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 3.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 8.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 5.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |