Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
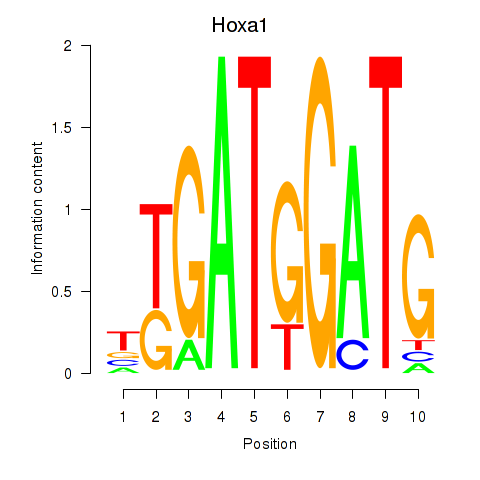
Results for Hoxa1
Z-value: 0.49
Transcription factors associated with Hoxa1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa1
|
ENSMUSG00000029844.9 | homeobox A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa1 | mm10_v2_chr6_-_52158292_52158324 | 0.33 | 5.1e-02 | Click! |
Activity profile of Hoxa1 motif
Sorted Z-values of Hoxa1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_12340723 | 1.97 |
ENSMUST00000168193.1
ENSMUST00000110616.1 ENSMUST00000064204.7 |
Actn2
|
actinin alpha 2 |
| chr11_-_43601540 | 1.96 |
ENSMUST00000020672.4
|
Fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr17_+_25298389 | 1.75 |
ENSMUST00000037453.2
|
Prss34
|
protease, serine, 34 |
| chr15_-_95528702 | 1.70 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chr11_+_24080664 | 0.99 |
ENSMUST00000118955.1
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr14_-_69284982 | 0.96 |
ENSMUST00000183882.1
ENSMUST00000037064.4 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr10_+_88885992 | 0.75 |
ENSMUST00000020255.6
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chrX_+_56454871 | 0.74 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr1_-_119053339 | 0.70 |
ENSMUST00000161301.1
ENSMUST00000161451.1 ENSMUST00000162607.1 |
Gli2
|
GLI-Kruppel family member GLI2 |
| chr3_+_114030532 | 0.70 |
ENSMUST00000123619.1
ENSMUST00000092155.5 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr7_-_30534180 | 0.66 |
ENSMUST00000044338.4
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr9_+_65890237 | 0.65 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr4_-_143299498 | 0.64 |
ENSMUST00000030317.7
|
Pdpn
|
podoplanin |
| chr14_-_69503316 | 0.62 |
ENSMUST00000179116.2
|
Gm21464
|
predicted gene, 21464 |
| chr7_+_73391160 | 0.62 |
ENSMUST00000128471.1
|
Rgma
|
RGM domain family, member A |
| chr1_-_45503282 | 0.59 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr2_-_131042682 | 0.58 |
ENSMUST00000028787.5
ENSMUST00000110239.1 ENSMUST00000110234.1 |
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4 |
| chr8_-_8639363 | 0.56 |
ENSMUST00000152698.1
|
Efnb2
|
ephrin B2 |
| chr18_-_13972617 | 0.52 |
ENSMUST00000025288.7
|
Zfp521
|
zinc finger protein 521 |
| chr9_-_21312255 | 0.52 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr15_+_102990576 | 0.50 |
ENSMUST00000001703.6
|
Hoxc8
|
homeobox C8 |
| chr2_+_165595009 | 0.46 |
ENSMUST00000088132.6
|
Eya2
|
eyes absent 2 homolog (Drosophila) |
| chr10_+_14523062 | 0.45 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr19_-_41743665 | 0.42 |
ENSMUST00000025993.3
|
Slit1
|
slit homolog 1 (Drosophila) |
| chr13_+_72628802 | 0.41 |
ENSMUST00000074372.4
|
Irx2
|
Iroquois related homeobox 2 (Drosophila) |
| chr6_+_142298419 | 0.40 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr7_+_144838590 | 0.37 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chr9_+_110344185 | 0.35 |
ENSMUST00000142100.1
|
Scap
|
SREBF chaperone |
| chr1_-_119053619 | 0.34 |
ENSMUST00000062483.8
|
Gli2
|
GLI-Kruppel family member GLI2 |
| chr2_+_32587057 | 0.32 |
ENSMUST00000102818.4
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr15_+_82252397 | 0.32 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr2_-_160619971 | 0.32 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr5_+_27261916 | 0.32 |
ENSMUST00000101471.3
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr4_-_143299463 | 0.31 |
ENSMUST00000119654.1
|
Pdpn
|
podoplanin |
| chr2_-_131043088 | 0.31 |
ENSMUST00000110240.3
ENSMUST00000066958.4 ENSMUST00000110235.1 |
Gfra4
|
glial cell line derived neurotrophic factor family receptor alpha 4 |
| chr17_-_51826562 | 0.30 |
ENSMUST00000024720.4
ENSMUST00000129667.1 ENSMUST00000156051.1 ENSMUST00000169480.1 ENSMUST00000148559.1 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr13_+_31806627 | 0.30 |
ENSMUST00000062292.2
|
Foxc1
|
forkhead box C1 |
| chr2_+_163225363 | 0.30 |
ENSMUST00000099110.3
ENSMUST00000165937.1 |
Tox2
|
TOX high mobility group box family member 2 |
| chrX_-_150813637 | 0.30 |
ENSMUST00000112700.1
|
Maged2
|
melanoma antigen, family D, 2 |
| chr9_-_96862903 | 0.28 |
ENSMUST00000121077.1
ENSMUST00000124923.1 |
Acpl2
|
acid phosphatase-like 2 |
| chrX_-_150812932 | 0.27 |
ENSMUST00000131241.1
ENSMUST00000147152.1 ENSMUST00000143843.1 |
Maged2
|
melanoma antigen, family D, 2 |
| chr5_-_145201829 | 0.26 |
ENSMUST00000162220.1
ENSMUST00000031632.8 |
Zkscan14
|
zinc finger with KRAB and SCAN domains 14 |
| chr19_+_46396885 | 0.26 |
ENSMUST00000039922.6
ENSMUST00000111867.2 ENSMUST00000120778.1 |
Sufu
|
suppressor of fused homolog (Drosophila) |
| chr9_+_58129321 | 0.25 |
ENSMUST00000034880.3
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr9_+_58129062 | 0.24 |
ENSMUST00000085677.2
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr10_+_79988584 | 0.24 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chr3_+_55782500 | 0.21 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr10_+_94576254 | 0.21 |
ENSMUST00000117929.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr4_+_119814495 | 0.21 |
ENSMUST00000106307.2
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr3_+_58526303 | 0.20 |
ENSMUST00000138848.1
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr2_-_152951688 | 0.20 |
ENSMUST00000109811.3
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr14_+_66344369 | 0.19 |
ENSMUST00000118426.1
ENSMUST00000121955.1 ENSMUST00000120229.1 ENSMUST00000134440.1 |
Stmn4
|
stathmin-like 4 |
| chr9_-_31464238 | 0.19 |
ENSMUST00000048050.7
|
Tmem45b
|
transmembrane protein 45b |
| chr4_+_101507947 | 0.18 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_+_138775735 | 0.17 |
ENSMUST00000030528.2
|
Pla2g2d
|
phospholipase A2, group IID |
| chr3_+_58525821 | 0.17 |
ENSMUST00000029387.8
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr3_+_68572245 | 0.17 |
ENSMUST00000170788.2
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_-_145315143 | 0.17 |
ENSMUST00000030339.6
|
Tnfrsf8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr5_+_137030275 | 0.17 |
ENSMUST00000041543.8
|
Vgf
|
VGF nerve growth factor inducible |
| chr2_+_90885860 | 0.16 |
ENSMUST00000111466.2
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr1_-_180813534 | 0.15 |
ENSMUST00000159789.1
ENSMUST00000081026.4 |
H3f3a
|
H3 histone, family 3A |
| chr1_+_153891646 | 0.15 |
ENSMUST00000050660.4
|
Teddm1
|
transmembrane epididymal protein 1 |
| chrX_-_75578188 | 0.15 |
ENSMUST00000033545.5
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr14_+_66344296 | 0.15 |
ENSMUST00000152093.1
ENSMUST00000074523.6 |
Stmn4
|
stathmin-like 4 |
| chr17_-_28622419 | 0.15 |
ENSMUST00000114767.1
|
Srpk1
|
serine/arginine-rich protein specific kinase 1 |
| chr11_+_58665561 | 0.15 |
ENSMUST00000072030.3
|
Olfr322
|
olfactory receptor 322 |
| chr16_+_36308045 | 0.15 |
ENSMUST00000114851.1
|
Gm4758
|
predicted gene 4758 |
| chr1_-_180813591 | 0.14 |
ENSMUST00000162118.1
ENSMUST00000159685.1 ENSMUST00000161308.1 |
H3f3a
|
H3 histone, family 3A |
| chr1_+_89454769 | 0.14 |
ENSMUST00000027521.8
ENSMUST00000074945.5 |
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr2_+_65845767 | 0.13 |
ENSMUST00000122912.1
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr17_+_78491549 | 0.12 |
ENSMUST00000079363.4
|
Gm10093
|
predicted pseudogene 10093 |
| chr4_+_101507855 | 0.12 |
ENSMUST00000038207.5
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr6_-_52012476 | 0.11 |
ENSMUST00000078214.5
|
Skap2
|
src family associated phosphoprotein 2 |
| chr14_-_103844173 | 0.11 |
ENSMUST00000022718.3
|
Ednrb
|
endothelin receptor type B |
| chr15_+_98167806 | 0.11 |
ENSMUST00000031914.4
|
AI836003
|
expressed sequence AI836003 |
| chr11_-_5898771 | 0.11 |
ENSMUST00000102921.3
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr10_-_10429839 | 0.11 |
ENSMUST00000045328.7
|
Adgb
|
androglobin |
| chr1_+_153665274 | 0.10 |
ENSMUST00000152114.1
ENSMUST00000111812.1 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr6_+_49319274 | 0.10 |
ENSMUST00000055559.7
ENSMUST00000114491.1 |
Ccdc126
|
coiled-coil domain containing 126 |
| chr6_+_116506516 | 0.10 |
ENSMUST00000075756.2
|
Olfr212
|
olfactory receptor 212 |
| chr13_+_99184733 | 0.10 |
ENSMUST00000056558.8
|
Zfp366
|
zinc finger protein 366 |
| chr13_-_103764502 | 0.09 |
ENSMUST00000074616.5
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr11_-_69549108 | 0.09 |
ENSMUST00000108659.1
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr8_-_105484350 | 0.09 |
ENSMUST00000044286.5
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr16_+_18877037 | 0.09 |
ENSMUST00000120532.1
ENSMUST00000004222.7 |
Hira
|
histone cell cycle regulation defective homolog A (S. cerevisiae) |
| chr9_-_4796218 | 0.09 |
ENSMUST00000027020.6
ENSMUST00000063508.7 ENSMUST00000163309.1 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr7_+_103979176 | 0.09 |
ENSMUST00000098185.1
|
Olfr635
|
olfactory receptor 635 |
| chr2_+_36452587 | 0.09 |
ENSMUST00000072854.1
|
Olfr340
|
olfactory receptor 340 |
| chr2_+_36230426 | 0.08 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_113930934 | 0.08 |
ENSMUST00000084885.5
ENSMUST00000009885.7 |
Ubp1
|
upstream binding protein 1 |
| chr4_+_150997081 | 0.07 |
ENSMUST00000030803.1
|
Uts2
|
urotensin 2 |
| chr10_+_90071095 | 0.07 |
ENSMUST00000183109.1
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr14_+_70077375 | 0.06 |
ENSMUST00000035908.1
|
Egr3
|
early growth response 3 |
| chr11_+_96351632 | 0.06 |
ENSMUST00000100523.5
|
Hoxb2
|
homeobox B2 |
| chr2_-_26246707 | 0.06 |
ENSMUST00000166349.1
|
C030048H21Rik
|
RIKEN cDNA C030048H21 gene |
| chr12_-_80260356 | 0.05 |
ENSMUST00000021554.8
|
Actn1
|
actinin, alpha 1 |
| chr3_+_138352378 | 0.05 |
ENSMUST00000090166.4
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr5_-_107289561 | 0.05 |
ENSMUST00000031224.8
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr15_-_78388757 | 0.05 |
ENSMUST00000169575.1
|
Tex33
|
testis expressed 33 |
| chr9_+_58129476 | 0.05 |
ENSMUST00000133287.1
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr10_+_81628540 | 0.04 |
ENSMUST00000123896.1
|
Ankrd24
|
ankyrin repeat domain 24 |
| chr12_+_53248677 | 0.04 |
ENSMUST00000101432.2
|
Npas3
|
neuronal PAS domain protein 3 |
| chr5_+_43515513 | 0.04 |
ENSMUST00000167522.1
ENSMUST00000144558.1 ENSMUST00000076939.6 |
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr15_+_74516196 | 0.04 |
ENSMUST00000042035.9
|
Bai1
|
brain-specific angiogenesis inhibitor 1 |
| chr17_-_28622479 | 0.04 |
ENSMUST00000130643.1
|
Srpk1
|
serine/arginine-rich protein specific kinase 1 |
| chr2_+_152962485 | 0.04 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr11_-_65159890 | 0.03 |
ENSMUST00000020855.3
ENSMUST00000108696.1 |
1700086D15Rik
|
RIKEN cDNA 1700086D15 gene |
| chr6_+_40442863 | 0.02 |
ENSMUST00000038907.8
ENSMUST00000141490.1 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chr7_+_140322602 | 0.02 |
ENSMUST00000078103.2
|
Olfr525
|
olfactory receptor 525 |
| chr2_+_124610573 | 0.02 |
ENSMUST00000103239.3
ENSMUST00000103240.2 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr17_+_70522149 | 0.02 |
ENSMUST00000140728.1
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr9_-_20336094 | 0.02 |
ENSMUST00000086473.3
|
Olfr18
|
olfactory receptor 18 |
| chr14_+_56575603 | 0.02 |
ENSMUST00000161553.1
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr7_-_16917184 | 0.01 |
ENSMUST00000173139.1
|
Calm3
|
calmodulin 3 |
| chr4_-_11981265 | 0.01 |
ENSMUST00000098260.2
|
Gm10604
|
predicted gene 10604 |
| chr4_-_108406676 | 0.01 |
ENSMUST00000184609.1
|
Gpx7
|
glutathione peroxidase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.0 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.3 | 1.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 1.0 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.1 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 1.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.3 | GO:1990869 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.4 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 0.3 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.6 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.5 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.4 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 2.0 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.6 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 1.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 2.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 0.9 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 2.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |