Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
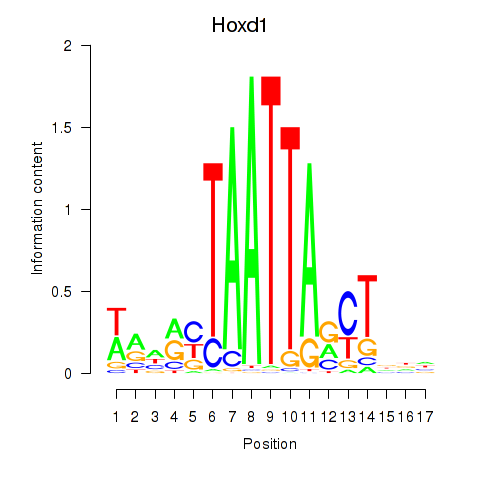
Results for Hoxd1
Z-value: 0.58
Transcription factors associated with Hoxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd1
|
ENSMUSG00000042448.4 | homeobox D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd1 | mm10_v2_chr2_+_74762980_74762980 | 0.07 | 7.0e-01 | Click! |
Activity profile of Hoxd1 motif
Sorted Z-values of Hoxd1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7966000 | 2.05 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr2_-_164638789 | 1.25 |
ENSMUST00000109336.1
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chr7_-_48843663 | 1.23 |
ENSMUST00000167786.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr7_-_140154712 | 1.19 |
ENSMUST00000059241.7
|
Sprn
|
shadow of prion protein |
| chr9_-_15301555 | 1.12 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr3_-_113291449 | 1.07 |
ENSMUST00000179568.1
|
Amy2a4
|
amylase 2a4 |
| chr17_-_45659312 | 1.06 |
ENSMUST00000120717.1
|
Capn11
|
calpain 11 |
| chr1_+_88070765 | 1.03 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr10_+_63024315 | 1.01 |
ENSMUST00000124784.1
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chrX_-_75843185 | 0.97 |
ENSMUST00000137192.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr2_+_109917639 | 0.93 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr2_+_69897255 | 0.90 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chrX_-_75843063 | 0.86 |
ENSMUST00000114057.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr4_+_102570065 | 0.85 |
ENSMUST00000097950.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr9_-_107872403 | 0.83 |
ENSMUST00000183035.1
|
Rbm6
|
RNA binding motif protein 6 |
| chr17_-_36032682 | 0.82 |
ENSMUST00000102678.4
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr19_+_58759700 | 0.82 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr1_+_88055467 | 0.76 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chrM_+_9870 | 0.73 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr12_-_98577940 | 0.73 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr18_-_60273267 | 0.73 |
ENSMUST00000090260.4
|
Gm4841
|
predicted gene 4841 |
| chr1_+_88055377 | 0.72 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_+_116067213 | 0.71 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr5_-_62766153 | 0.68 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_+_66797039 | 0.67 |
ENSMUST00000098612.2
|
Gm10647
|
predicted gene 10647 |
| chr2_-_155074447 | 0.67 |
ENSMUST00000137242.1
ENSMUST00000054607.9 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr1_-_65186456 | 0.66 |
ENSMUST00000169032.1
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr2_+_69897220 | 0.66 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_-_20943413 | 0.65 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr1_-_24612700 | 0.64 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr18_-_3281036 | 0.61 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr17_-_28560704 | 0.59 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr19_+_3323301 | 0.58 |
ENSMUST00000025835.4
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr6_-_130193112 | 0.58 |
ENSMUST00000112032.1
ENSMUST00000071554.2 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr2_-_156392829 | 0.58 |
ENSMUST00000088578.2
|
2900097C17Rik
|
RIKEN cDNA 2900097C17 gene |
| chr3_+_19985612 | 0.58 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr5_+_90561102 | 0.58 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr6_+_149582012 | 0.57 |
ENSMUST00000144085.2
|
Gm21814
|
predicted gene, 21814 |
| chrM_+_10167 | 0.54 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr5_-_77115145 | 0.53 |
ENSMUST00000081964.5
|
Hopx
|
HOP homeobox |
| chr6_-_83656082 | 0.53 |
ENSMUST00000014686.2
|
Clec4f
|
C-type lectin domain family 4, member f |
| chr15_+_98571004 | 0.53 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr2_+_126556128 | 0.50 |
ENSMUST00000141482.2
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr7_+_79273201 | 0.48 |
ENSMUST00000037315.6
|
Abhd2
|
abhydrolase domain containing 2 |
| chr9_-_45204083 | 0.48 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr6_-_130026954 | 0.46 |
ENSMUST00000074056.2
|
Klra6
|
killer cell lectin-like receptor, subfamily A, member 6 |
| chr5_-_87490869 | 0.46 |
ENSMUST00000147854.1
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr1_-_4360256 | 0.45 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chrX_-_143933089 | 0.44 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr9_+_72958785 | 0.44 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr15_+_25773985 | 0.43 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr8_-_41041828 | 0.43 |
ENSMUST00000051379.7
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr5_-_123012874 | 0.43 |
ENSMUST00000172729.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr9_-_123851855 | 0.43 |
ENSMUST00000184082.1
ENSMUST00000167595.2 |
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chrX_+_107255878 | 0.43 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr6_-_57535422 | 0.43 |
ENSMUST00000042766.3
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr3_-_116712644 | 0.42 |
ENSMUST00000029569.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr5_+_29195983 | 0.41 |
ENSMUST00000160888.1
ENSMUST00000159272.1 ENSMUST00000001247.5 ENSMUST00000161398.1 ENSMUST00000160246.1 |
Rnf32
|
ring finger protein 32 |
| chr6_-_130129898 | 0.41 |
ENSMUST00000014476.5
|
Klra8
|
killer cell lectin-like receptor, subfamily A, member 8 |
| chr5_-_63968867 | 0.41 |
ENSMUST00000154169.1
|
Rell1
|
RELT-like 1 |
| chr11_-_121388186 | 0.40 |
ENSMUST00000106107.2
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr11_+_109543694 | 0.39 |
ENSMUST00000106696.1
|
Arsg
|
arylsulfatase G |
| chr2_+_121956651 | 0.39 |
ENSMUST00000110574.1
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr9_-_85749308 | 0.39 |
ENSMUST00000039213.8
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr13_-_67332525 | 0.38 |
ENSMUST00000168892.1
ENSMUST00000109735.2 |
Zfp595
|
zinc finger protein 595 |
| chr4_+_147492417 | 0.38 |
ENSMUST00000105721.2
|
Gm13152
|
predicted gene 13152 |
| chr2_-_69712461 | 0.37 |
ENSMUST00000102706.3
ENSMUST00000073152.6 |
Fastkd1
|
FAST kinase domains 1 |
| chr14_+_55559993 | 0.37 |
ENSMUST00000117236.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_-_85722474 | 0.37 |
ENSMUST00000119077.1
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr5_-_108795352 | 0.37 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr9_+_119341487 | 0.36 |
ENSMUST00000175743.1
ENSMUST00000176397.1 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr16_+_11406618 | 0.36 |
ENSMUST00000122168.1
|
Snx29
|
sorting nexin 29 |
| chr6_+_78370877 | 0.36 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr7_+_29816061 | 0.35 |
ENSMUST00000032796.6
ENSMUST00000178162.1 |
Zfp790
|
zinc finger protein 790 |
| chr16_-_16600533 | 0.35 |
ENSMUST00000159542.1
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_+_119341294 | 0.35 |
ENSMUST00000039784.5
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr8_+_46010596 | 0.34 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chr2_-_160619971 | 0.34 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr3_-_16006332 | 0.33 |
ENSMUST00000108347.2
|
Gm5150
|
predicted gene 5150 |
| chr15_-_101892916 | 0.33 |
ENSMUST00000100179.1
|
Krt76
|
keratin 76 |
| chr17_+_88440711 | 0.33 |
ENSMUST00000112238.2
ENSMUST00000155640.1 |
Foxn2
|
forkhead box N2 |
| chr2_+_121956411 | 0.33 |
ENSMUST00000110578.1
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr12_+_10390756 | 0.33 |
ENSMUST00000020947.5
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr15_-_8710734 | 0.32 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr18_+_12741324 | 0.31 |
ENSMUST00000115857.2
ENSMUST00000121018.1 ENSMUST00000119108.1 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr14_+_47298260 | 0.31 |
ENSMUST00000166743.1
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr2_+_91257323 | 0.31 |
ENSMUST00000111349.2
ENSMUST00000131711.1 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_-_40242285 | 0.31 |
ENSMUST00000026499.5
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr9_-_55919605 | 0.31 |
ENSMUST00000037408.8
|
Scaper
|
S phase cyclin A-associated protein in the ER |
| chr3_-_15426427 | 0.31 |
ENSMUST00000099201.3
|
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr2_+_32606946 | 0.30 |
ENSMUST00000113290.1
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chrX_-_60893430 | 0.30 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr19_-_37330613 | 0.30 |
ENSMUST00000131070.1
|
Ide
|
insulin degrading enzyme |
| chr19_-_39649046 | 0.30 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chrX_-_99470672 | 0.30 |
ENSMUST00000113797.3
ENSMUST00000113790.1 ENSMUST00000036354.6 ENSMUST00000167246.1 |
Pja1
|
praja1, RING-H2 motif containing |
| chr6_-_69284319 | 0.29 |
ENSMUST00000103349.1
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr2_-_177267036 | 0.29 |
ENSMUST00000108963.1
|
Gm14409
|
predicted gene 14409 |
| chr2_-_116067391 | 0.28 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr2_+_32606979 | 0.28 |
ENSMUST00000113289.1
ENSMUST00000095044.3 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr10_-_8886033 | 0.28 |
ENSMUST00000015449.5
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr6_+_68161415 | 0.28 |
ENSMUST00000168090.1
|
Igkv1-115
|
immunoglobulin kappa variable 1-115 |
| chr13_-_102905740 | 0.27 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_102768771 | 0.26 |
ENSMUST00000112852.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_-_33814516 | 0.26 |
ENSMUST00000044455.5
ENSMUST00000115167.1 |
Zfp451
|
zinc finger protein 451 |
| chr10_+_94575257 | 0.25 |
ENSMUST00000121471.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr13_+_42866247 | 0.25 |
ENSMUST00000131942.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr4_+_95557494 | 0.25 |
ENSMUST00000079223.4
ENSMUST00000177394.1 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr6_-_116716888 | 0.24 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr2_+_3424123 | 0.24 |
ENSMUST00000061852.5
ENSMUST00000100463.3 ENSMUST00000102988.3 ENSMUST00000115066.1 |
Dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr1_-_33814591 | 0.23 |
ENSMUST00000019861.6
|
Zfp451
|
zinc finger protein 451 |
| chr6_-_69400097 | 0.23 |
ENSMUST00000177795.1
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr1_+_65186727 | 0.23 |
ENSMUST00000097707.4
ENSMUST00000081154.7 |
Pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr12_-_112802646 | 0.23 |
ENSMUST00000124526.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr6_-_124779686 | 0.22 |
ENSMUST00000147669.1
ENSMUST00000128697.1 ENSMUST00000032218.3 ENSMUST00000112475.2 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr3_-_15848419 | 0.22 |
ENSMUST00000108354.1
ENSMUST00000108349.1 ENSMUST00000108352.2 ENSMUST00000108350.1 ENSMUST00000050623.4 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr11_-_107337556 | 0.21 |
ENSMUST00000040380.6
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chrX_+_16619698 | 0.21 |
ENSMUST00000026013.5
|
Maoa
|
monoamine oxidase A |
| chr12_+_80644212 | 0.20 |
ENSMUST00000085245.5
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr8_+_113635787 | 0.20 |
ENSMUST00000035777.8
|
Mon1b
|
MON1 homolog b (yeast) |
| chr10_-_86011833 | 0.20 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr10_-_92375367 | 0.19 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr2_-_150255591 | 0.19 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr7_+_30977043 | 0.19 |
ENSMUST00000058093.4
|
Fam187b
|
family with sequence similarity 187, member B |
| chr3_-_41742471 | 0.19 |
ENSMUST00000026866.8
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr5_+_107497762 | 0.19 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr13_+_24802020 | 0.19 |
ENSMUST00000155575.1
|
BC005537
|
cDNA sequence BC005537 |
| chrX_-_160138375 | 0.18 |
ENSMUST00000033662.8
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr11_-_99374895 | 0.18 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr13_-_102906046 | 0.17 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_48446128 | 0.17 |
ENSMUST00000124720.1
|
Tmem260
|
transmembrane protein 260 |
| chr4_+_103143052 | 0.17 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr8_+_113635550 | 0.16 |
ENSMUST00000179926.1
|
Mon1b
|
MON1 homolog b (yeast) |
| chr6_+_115931922 | 0.16 |
ENSMUST00000032471.6
|
Rho
|
rhodopsin |
| chr3_-_96452306 | 0.16 |
ENSMUST00000093126.4
ENSMUST00000098841.3 |
BC107364
|
cDNA sequence BC107364 |
| chr1_+_157506777 | 0.16 |
ENSMUST00000027881.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr4_-_148152059 | 0.16 |
ENSMUST00000056965.5
ENSMUST00000168503.1 ENSMUST00000152098.1 |
Fbxo6
|
F-box protein 6 |
| chrX_+_160768013 | 0.16 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr1_+_157506728 | 0.16 |
ENSMUST00000086130.2
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr14_+_69347587 | 0.16 |
ENSMUST00000064831.5
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr14_-_51071442 | 0.16 |
ENSMUST00000048478.5
|
Olfr750
|
olfactory receptor 750 |
| chr5_-_90640464 | 0.15 |
ENSMUST00000031317.6
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr14_+_122034660 | 0.15 |
ENSMUST00000045976.6
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr11_+_99873389 | 0.15 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chrX_-_143933204 | 0.14 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr7_+_135268579 | 0.14 |
ENSMUST00000097983.3
|
Nps
|
neuropeptide S |
| chr18_+_31609512 | 0.14 |
ENSMUST00000164667.1
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr10_+_57650990 | 0.14 |
ENSMUST00000175852.1
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr19_-_41933276 | 0.14 |
ENSMUST00000075280.4
ENSMUST00000112123.2 |
Exosc1
|
exosome component 1 |
| chr15_-_37459327 | 0.14 |
ENSMUST00000119730.1
ENSMUST00000120746.1 |
Ncald
|
neurocalcin delta |
| chr6_+_116650674 | 0.14 |
ENSMUST00000067354.5
ENSMUST00000178241.1 |
8430408G22Rik
|
RIKEN cDNA 8430408G22 gene |
| chr7_-_24724237 | 0.13 |
ENSMUST00000081657.4
|
Gm4763
|
predicted gene 4763 |
| chr13_+_24801657 | 0.13 |
ENSMUST00000019276.4
|
BC005537
|
cDNA sequence BC005537 |
| chr2_+_170731807 | 0.13 |
ENSMUST00000029075.4
|
Dok5
|
docking protein 5 |
| chr13_-_74807913 | 0.12 |
ENSMUST00000065629.4
|
Cast
|
calpastatin |
| chr5_+_145204523 | 0.12 |
ENSMUST00000085671.3
ENSMUST00000031601.7 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr2_+_37516618 | 0.12 |
ENSMUST00000065441.6
|
Gpr21
|
G protein-coupled receptor 21 |
| chr16_+_18836573 | 0.12 |
ENSMUST00000055413.6
|
2510002D24Rik
|
RIKEN cDNA 2510002D24 gene |
| chr4_+_145514884 | 0.12 |
ENSMUST00000105741.1
|
Gm13225
|
predicted gene 13225 |
| chr17_-_32822200 | 0.12 |
ENSMUST00000179695.1
|
Zfp799
|
zinc finger protein 799 |
| chr10_-_112928974 | 0.11 |
ENSMUST00000099276.2
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr7_+_130774069 | 0.11 |
ENSMUST00000048453.5
|
Btbd16
|
BTB (POZ) domain containing 16 |
| chr11_-_100088226 | 0.11 |
ENSMUST00000107419.1
|
Krt32
|
keratin 32 |
| chr3_+_94837702 | 0.10 |
ENSMUST00000107266.1
ENSMUST00000042402.5 ENSMUST00000107269.1 |
Pogz
|
pogo transposable element with ZNF domain |
| chr15_+_38661904 | 0.10 |
ENSMUST00000022904.6
|
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr15_-_8710409 | 0.10 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_-_32886083 | 0.10 |
ENSMUST00000178401.1
|
Zfp870
|
zinc finger protein 870 |
| chr11_-_99244058 | 0.10 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr12_-_84617326 | 0.10 |
ENSMUST00000021666.4
|
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr2_-_132247747 | 0.09 |
ENSMUST00000110163.1
ENSMUST00000180286.1 ENSMUST00000028816.2 |
Tmem230
|
transmembrane protein 230 |
| chr11_-_30649510 | 0.09 |
ENSMUST00000074613.3
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr18_+_37355271 | 0.09 |
ENSMUST00000051163.1
|
Pcdhb8
|
protocadherin beta 8 |
| chr17_-_36462329 | 0.09 |
ENSMUST00000169950.1
ENSMUST00000057502.7 |
H2-M10.4
|
histocompatibility 2, M region locus 10.4 |
| chr5_-_3647806 | 0.09 |
ENSMUST00000119783.1
ENSMUST00000007559.8 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr15_+_82256023 | 0.09 |
ENSMUST00000143238.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr19_+_23723279 | 0.09 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr8_-_122915987 | 0.09 |
ENSMUST00000098333.4
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr19_-_53944621 | 0.08 |
ENSMUST00000135402.2
|
Bbip1
|
BBSome interacting protein 1 |
| chr4_+_146514920 | 0.08 |
ENSMUST00000140089.1
ENSMUST00000179175.1 |
Gm13247
|
predicted gene 13247 |
| chr9_-_105395237 | 0.08 |
ENSMUST00000140851.1
|
Nek11
|
NIMA (never in mitosis gene a)-related expressed kinase 11 |
| chr7_+_18245347 | 0.08 |
ENSMUST00000066780.4
|
Mill1
|
MHC I like leukocyte 1 |
| chr16_+_45224315 | 0.08 |
ENSMUST00000102802.3
ENSMUST00000063654.4 |
Btla
|
B and T lymphocyte associated |
| chr7_+_127511976 | 0.08 |
ENSMUST00000098025.4
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr19_-_32196393 | 0.08 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr1_-_185329331 | 0.08 |
ENSMUST00000027921.4
ENSMUST00000110975.1 ENSMUST00000110974.3 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr9_-_86880647 | 0.08 |
ENSMUST00000167014.1
|
Snap91
|
synaptosomal-associated protein 91 |
| chr6_-_3399545 | 0.07 |
ENSMUST00000120087.3
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr2_-_84715160 | 0.07 |
ENSMUST00000035840.5
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr2_-_45117349 | 0.07 |
ENSMUST00000176438.2
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr16_+_88728828 | 0.06 |
ENSMUST00000060494.6
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr11_-_4095344 | 0.06 |
ENSMUST00000004868.5
|
Mtfp1
|
mitochondrial fission process 1 |
| chr17_+_34931253 | 0.06 |
ENSMUST00000007253.5
|
Neu1
|
neuraminidase 1 |
| chr9_-_70934808 | 0.06 |
ENSMUST00000034731.8
|
Lipc
|
lipase, hepatic |
| chr1_+_88306731 | 0.06 |
ENSMUST00000040210.7
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr13_-_105054895 | 0.06 |
ENSMUST00000063551.5
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein |
| chr9_+_105395348 | 0.06 |
ENSMUST00000035181.3
|
Aste1
|
asteroid homolog 1 (Drosophila) |
| chr1_-_158356258 | 0.06 |
ENSMUST00000004133.8
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr19_-_39886730 | 0.05 |
ENSMUST00000168838.1
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr4_-_14621669 | 0.05 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_+_57650977 | 0.05 |
ENSMUST00000066028.6
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr11_-_115419917 | 0.05 |
ENSMUST00000106537.1
ENSMUST00000043931.2 ENSMUST00000073791.3 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1903918 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.4 | 1.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 0.9 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 2.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.8 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.2 | 0.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 0.7 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.2 | 0.5 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 2.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.4 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 1.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.9 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.0 | 0.4 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.5 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.2 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0098830 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0008410 | acetyl-CoA C-acetyltransferase activity(GO:0003985) CoA-transferase activity(GO:0008410) palmitoyl-CoA oxidase activity(GO:0016401) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 3.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |