Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hsf2
Z-value: 0.72
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSMUSG00000019878.7 | heat shock factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | mm10_v2_chr10_+_57486354_57486414 | 0.17 | 3.1e-01 | Click! |
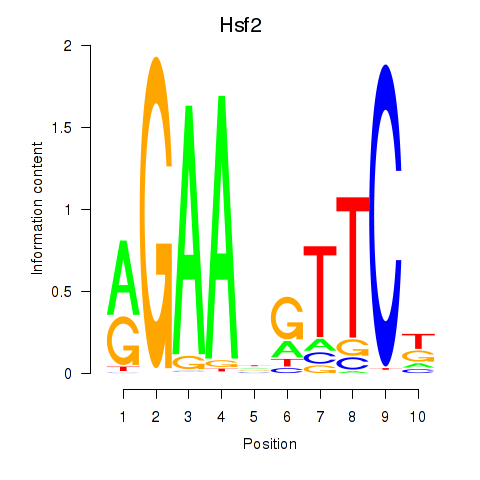
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_66935333 | 2.01 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr4_-_141825997 | 1.33 |
ENSMUST00000102481.3
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr9_+_8544196 | 1.28 |
ENSMUST00000050433.6
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chrX_+_21484532 | 1.24 |
ENSMUST00000089188.2
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr11_-_100121558 | 1.09 |
ENSMUST00000007275.2
|
Krt13
|
keratin 13 |
| chr9_+_98490522 | 1.05 |
ENSMUST00000035029.2
|
Rbp2
|
retinol binding protein 2, cellular |
| chr13_-_37049203 | 1.00 |
ENSMUST00000037491.8
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr6_+_41521782 | 0.91 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr16_-_36455378 | 0.81 |
ENSMUST00000068182.2
|
Stfa3
|
stefin A3 |
| chr17_-_31129602 | 0.81 |
ENSMUST00000024827.4
|
Tff3
|
trefoil factor 3, intestinal |
| chr16_-_36367623 | 0.80 |
ENSMUST00000096089.2
|
BC100530
|
cDNA sequence BC100530 |
| chr16_+_17980565 | 0.80 |
ENSMUST00000075371.3
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr5_+_76656512 | 0.80 |
ENSMUST00000086909.4
|
Gm10430
|
predicted gene 10430 |
| chrX_+_164090187 | 0.78 |
ENSMUST00000015545.3
|
Tmem27
|
transmembrane protein 27 |
| chr5_+_135887905 | 0.73 |
ENSMUST00000005077.6
|
Hspb1
|
heat shock protein 1 |
| chr16_+_36156801 | 0.72 |
ENSMUST00000079184.4
|
Stfa2l1
|
stefin A2 like 1 |
| chr19_+_58670358 | 0.68 |
ENSMUST00000057270.7
|
Pnlip
|
pancreatic lipase |
| chr7_+_127211608 | 0.67 |
ENSMUST00000032910.6
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_-_126463100 | 0.67 |
ENSMUST00000032974.6
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_+_40811089 | 0.64 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr16_-_36408349 | 0.64 |
ENSMUST00000023619.6
|
Stfa2
|
stefin A2 |
| chr18_+_36664060 | 0.63 |
ENSMUST00000036765.7
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr5_+_135887988 | 0.62 |
ENSMUST00000111155.1
|
Hspb1
|
heat shock protein 1 |
| chr2_+_43748802 | 0.62 |
ENSMUST00000112824.1
ENSMUST00000055776.7 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chrX_+_157702574 | 0.61 |
ENSMUST00000112520.1
|
Smpx
|
small muscle protein, X-linked |
| chr1_-_37496095 | 0.61 |
ENSMUST00000148047.1
ENSMUST00000143636.1 |
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr9_+_72438519 | 0.59 |
ENSMUST00000184604.1
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr16_+_36277145 | 0.59 |
ENSMUST00000042097.9
|
Stfa1
|
stefin A1 |
| chr6_-_78468863 | 0.59 |
ENSMUST00000032089.2
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr12_-_110696248 | 0.58 |
ENSMUST00000124156.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr7_+_16781341 | 0.58 |
ENSMUST00000108496.2
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr6_+_60944472 | 0.57 |
ENSMUST00000129603.1
|
Mmrn1
|
multimerin 1 |
| chr18_-_68429235 | 0.55 |
ENSMUST00000052347.6
|
Mc2r
|
melanocortin 2 receptor |
| chr6_-_102464667 | 0.54 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr14_-_70630149 | 0.52 |
ENSMUST00000022694.9
|
Dmtn
|
dematin actin binding protein |
| chr6_-_40951826 | 0.51 |
ENSMUST00000073642.5
|
Gm4744
|
predicted gene 4744 |
| chr5_+_3571664 | 0.50 |
ENSMUST00000008451.5
|
1700109H08Rik
|
RIKEN cDNA 1700109H08 gene |
| chr3_+_79345361 | 0.50 |
ENSMUST00000164216.1
|
Gm17359
|
predicted gene, 17359 |
| chr12_+_104101087 | 0.50 |
ENSMUST00000021495.3
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr1_+_134111233 | 0.50 |
ENSMUST00000159963.1
ENSMUST00000160060.1 |
Chit1
|
chitinase 1 (chitotriosidase) |
| chr1_-_55088156 | 0.49 |
ENSMUST00000127861.1
ENSMUST00000144077.1 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr13_+_75967704 | 0.49 |
ENSMUST00000022081.1
|
Spata9
|
spermatogenesis associated 9 |
| chr16_+_36210403 | 0.49 |
ENSMUST00000089628.3
|
Gm5416
|
predicted gene 5416 |
| chr1_+_166130467 | 0.48 |
ENSMUST00000166860.1
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr12_-_76795489 | 0.47 |
ENSMUST00000082431.3
|
Gpx2
|
glutathione peroxidase 2 |
| chr1_-_55088024 | 0.47 |
ENSMUST00000027123.8
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr1_-_193273099 | 0.46 |
ENSMUST00000009777.2
|
G0s2
|
G0/G1 switch gene 2 |
| chr15_+_62039216 | 0.46 |
ENSMUST00000183297.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr9_-_18512885 | 0.45 |
ENSMUST00000034653.6
|
Muc16
|
mucin 16 |
| chr3_-_106149761 | 0.45 |
ENSMUST00000149836.1
|
Chi3l3
|
chitinase 3-like 3 |
| chr16_-_16869255 | 0.45 |
ENSMUST00000075017.4
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr12_-_110696289 | 0.44 |
ENSMUST00000021698.6
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr1_+_136467958 | 0.44 |
ENSMUST00000047817.6
|
Kif14
|
kinesin family member 14 |
| chr16_-_19883873 | 0.44 |
ENSMUST00000100083.3
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr18_+_23415400 | 0.43 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr3_+_41024369 | 0.43 |
ENSMUST00000099121.3
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_+_166130238 | 0.42 |
ENSMUST00000060833.7
ENSMUST00000166159.1 |
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr5_-_5664196 | 0.42 |
ENSMUST00000061008.3
ENSMUST00000054865.6 |
A330021E22Rik
|
RIKEN cDNA A330021E22 gene |
| chr13_-_49248146 | 0.40 |
ENSMUST00000119721.1
ENSMUST00000058196.6 |
Susd3
|
sushi domain containing 3 |
| chr17_-_43543639 | 0.40 |
ENSMUST00000178772.1
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr10_+_28668360 | 0.40 |
ENSMUST00000060409.6
ENSMUST00000056097.4 ENSMUST00000105516.2 |
Themis
|
thymocyte selection associated |
| chr6_-_129275360 | 0.40 |
ENSMUST00000032259.3
|
Cd69
|
CD69 antigen |
| chr13_-_92794809 | 0.39 |
ENSMUST00000022213.7
|
Thbs4
|
thrombospondin 4 |
| chr16_-_55934845 | 0.39 |
ENSMUST00000121129.1
ENSMUST00000023270.7 |
Cep97
|
centrosomal protein 97 |
| chr5_-_73191848 | 0.39 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr2_+_29124106 | 0.39 |
ENSMUST00000129544.1
|
Setx
|
senataxin |
| chr2_-_154892782 | 0.38 |
ENSMUST00000166171.1
ENSMUST00000161172.1 |
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr6_-_125494754 | 0.38 |
ENSMUST00000032492.8
|
Cd9
|
CD9 antigen |
| chr4_-_117125618 | 0.38 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr11_-_89302545 | 0.38 |
ENSMUST00000061728.3
|
Nog
|
noggin |
| chr6_-_78378851 | 0.38 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr17_+_28530834 | 0.37 |
ENSMUST00000025060.2
|
Armc12
|
armadillo repeat containing 12 |
| chr1_+_40515362 | 0.37 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr8_+_92901387 | 0.37 |
ENSMUST00000104947.2
|
Capns2
|
calpain, small subunit 2 |
| chr2_-_154892887 | 0.36 |
ENSMUST00000099173.4
|
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr3_-_129755305 | 0.36 |
ENSMUST00000029653.2
|
Egf
|
epidermal growth factor |
| chr10_+_128225830 | 0.36 |
ENSMUST00000026455.7
|
Mip
|
major intrinsic protein of eye lens fiber |
| chr12_-_110696332 | 0.36 |
ENSMUST00000094361.4
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr11_+_116671658 | 0.36 |
ENSMUST00000106378.1
ENSMUST00000144049.1 |
1810032O08Rik
|
RIKEN cDNA 1810032O08 gene |
| chr17_-_13404191 | 0.35 |
ENSMUST00000115650.1
|
Gm8597
|
predicted gene 8597 |
| chr13_-_111490028 | 0.34 |
ENSMUST00000091236.4
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr13_+_44729794 | 0.34 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_-_3504766 | 0.34 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr16_+_19760232 | 0.34 |
ENSMUST00000079780.3
ENSMUST00000164397.1 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chrX_+_136033367 | 0.33 |
ENSMUST00000057625.2
|
Arxes1
|
adipocyte-related X-chromosome expressed sequence 1 |
| chr9_-_107985863 | 0.33 |
ENSMUST00000048568.4
|
Fam212a
|
family with sequence similarity 212, member A |
| chr9_+_108986163 | 0.33 |
ENSMUST00000052724.3
|
Ucn2
|
urocortin 2 |
| chr7_+_110772604 | 0.33 |
ENSMUST00000005829.6
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_-_120982260 | 0.32 |
ENSMUST00000033169.8
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr2_-_149798701 | 0.32 |
ENSMUST00000148202.1
ENSMUST00000139471.1 |
Gm14133
|
predicted gene 14133 |
| chr1_-_75133866 | 0.32 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr16_+_19760195 | 0.32 |
ENSMUST00000121344.1
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr10_+_130322845 | 0.32 |
ENSMUST00000042586.8
|
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr2_+_30061754 | 0.31 |
ENSMUST00000149578.1
ENSMUST00000102866.3 |
Set
|
SET nuclear oncogene |
| chr1_-_193035651 | 0.31 |
ENSMUST00000016344.7
|
Syt14
|
synaptotagmin XIV |
| chr5_-_38876693 | 0.31 |
ENSMUST00000169819.1
ENSMUST00000171633.1 |
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr7_+_44188205 | 0.31 |
ENSMUST00000073713.6
|
Klk1b24
|
kallikrein 1-related peptidase b24 |
| chr3_-_92458715 | 0.31 |
ENSMUST00000058142.3
|
Sprr3
|
small proline-rich protein 3 |
| chr1_-_45890078 | 0.30 |
ENSMUST00000183590.1
|
Gm5269
|
predicted gene 5269 |
| chr1_-_79858627 | 0.30 |
ENSMUST00000027467.4
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr13_+_38204928 | 0.29 |
ENSMUST00000091641.5
ENSMUST00000178564.1 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr10_-_88356990 | 0.29 |
ENSMUST00000020249.1
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr12_-_17324703 | 0.29 |
ENSMUST00000020884.9
ENSMUST00000095820.5 ENSMUST00000127185.1 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr1_-_136234113 | 0.29 |
ENSMUST00000120339.1
ENSMUST00000048668.8 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr10_-_62507737 | 0.29 |
ENSMUST00000020271.6
|
Srgn
|
serglycin |
| chr4_-_94928789 | 0.29 |
ENSMUST00000030309.5
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr5_-_123749371 | 0.29 |
ENSMUST00000182955.1
ENSMUST00000182489.1 ENSMUST00000050827.7 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr4_+_134102581 | 0.28 |
ENSMUST00000074690.4
ENSMUST00000070246.2 ENSMUST00000156750.1 |
Ubxn11
|
UBX domain protein 11 |
| chr14_-_65833963 | 0.28 |
ENSMUST00000022613.9
|
Esco2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae) |
| chr11_+_49513618 | 0.28 |
ENSMUST00000060434.2
|
Olfr1384
|
olfactory receptor 1384 |
| chr6_-_7692867 | 0.28 |
ENSMUST00000115542.1
ENSMUST00000148349.1 |
Asns
|
asparagine synthetase |
| chr11_+_76904475 | 0.28 |
ENSMUST00000142166.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr9_-_53975246 | 0.28 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr13_+_84222286 | 0.28 |
ENSMUST00000057495.8
|
Tmem161b
|
transmembrane protein 161B |
| chr1_+_186749368 | 0.28 |
ENSMUST00000180869.1
|
A430105J06Rik
|
RIKEN cDNA A430105J06 gene |
| chr14_-_63193541 | 0.28 |
ENSMUST00000038229.4
|
Neil2
|
nei like 2 (E. coli) |
| chr7_+_44216456 | 0.28 |
ENSMUST00000074359.2
|
Klk1b5
|
kallikrein 1-related peptidase b5 |
| chr12_-_110695860 | 0.28 |
ENSMUST00000149189.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr2_+_154200371 | 0.28 |
ENSMUST00000028987.6
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr15_-_101491509 | 0.27 |
ENSMUST00000023718.7
|
5430421N21Rik
|
RIKEN cDNA 5430421N21 gene |
| chr4_+_17853451 | 0.27 |
ENSMUST00000029881.3
|
Mmp16
|
matrix metallopeptidase 16 |
| chr1_-_144249134 | 0.27 |
ENSMUST00000172388.1
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr2_-_74579379 | 0.27 |
ENSMUST00000130586.1
|
Lnp
|
limb and neural patterns |
| chr4_+_108479081 | 0.27 |
ENSMUST00000155068.1
|
Zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr11_-_69878018 | 0.27 |
ENSMUST00000178597.1
|
Tmem95
|
transmembrane protein 95 |
| chr19_-_16780822 | 0.26 |
ENSMUST00000068156.6
|
Vps13a
|
vacuolar protein sorting 13A (yeast) |
| chrX_+_135993820 | 0.26 |
ENSMUST00000058119.7
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr13_+_20794119 | 0.26 |
ENSMUST00000021757.3
|
Aoah
|
acyloxyacyl hydrolase |
| chr19_-_10203880 | 0.26 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr6_-_7693184 | 0.26 |
ENSMUST00000031766.5
|
Asns
|
asparagine synthetase |
| chr19_-_5845471 | 0.26 |
ENSMUST00000174287.1
ENSMUST00000173672.1 |
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr13_+_76579670 | 0.26 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr19_-_34166037 | 0.26 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr5_-_123749393 | 0.26 |
ENSMUST00000057795.5
ENSMUST00000111515.1 ENSMUST00000182309.1 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chrX_-_48594373 | 0.26 |
ENSMUST00000088898.4
ENSMUST00000072292.5 |
Zfp280c
|
zinc finger protein 280C |
| chr16_-_55934797 | 0.26 |
ENSMUST00000122280.1
ENSMUST00000121703.2 |
Cep97
|
centrosomal protein 97 |
| chr1_-_176807124 | 0.26 |
ENSMUST00000057037.7
|
Cep170
|
centrosomal protein 170 |
| chr9_-_71592312 | 0.25 |
ENSMUST00000166112.1
|
Myzap
|
myocardial zonula adherens protein |
| chr6_-_41680723 | 0.25 |
ENSMUST00000031901.5
|
Trpv5
|
transient receptor potential cation channel, subfamily V, member 5 |
| chr17_+_34972687 | 0.25 |
ENSMUST00000007248.3
|
Hspa1l
|
heat shock protein 1-like |
| chr16_-_59553970 | 0.25 |
ENSMUST00000139989.1
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr6_+_51470339 | 0.25 |
ENSMUST00000094623.3
|
Cbx3
|
chromobox 3 |
| chr6_-_7693110 | 0.25 |
ENSMUST00000126303.1
|
Asns
|
asparagine synthetase |
| chr4_+_21848039 | 0.24 |
ENSMUST00000098238.2
ENSMUST00000108229.1 |
Sfrs18
|
serine/arginine-rich splicing factor 18 |
| chr11_-_69858687 | 0.24 |
ENSMUST00000125571.1
|
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr8_-_80739497 | 0.24 |
ENSMUST00000043359.8
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr7_+_142434977 | 0.24 |
ENSMUST00000118276.1
ENSMUST00000105976.1 ENSMUST00000097939.2 |
Syt8
|
synaptotagmin VIII |
| chr10_-_62422427 | 0.24 |
ENSMUST00000020277.8
|
Hkdc1
|
hexokinase domain containing 1 |
| chrX_-_19237841 | 0.24 |
ENSMUST00000180592.1
|
Gm26652
|
predicted gene, 26652 |
| chr9_-_77045788 | 0.24 |
ENSMUST00000034911.6
|
Tinag
|
tubulointerstitial nephritis antigen |
| chr7_-_4752972 | 0.24 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr17_+_47385386 | 0.24 |
ENSMUST00000024774.7
ENSMUST00000145462.1 |
Guca1b
|
guanylate cyclase activator 1B |
| chr2_+_35109482 | 0.24 |
ENSMUST00000028235.4
ENSMUST00000156933.1 ENSMUST00000028237.8 |
Cep110
|
centrosomal protein 110 |
| chr6_-_83536215 | 0.24 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_+_43950614 | 0.24 |
ENSMUST00000072204.4
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr13_+_44729535 | 0.24 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr6_-_136941887 | 0.24 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_+_103424120 | 0.24 |
ENSMUST00000171693.1
|
Elf5
|
E74-like factor 5 |
| chr2_+_144527718 | 0.23 |
ENSMUST00000028914.2
ENSMUST00000110017.2 |
Polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr5_+_32611171 | 0.23 |
ENSMUST00000072311.6
ENSMUST00000168707.2 |
Yes1
|
Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 |
| chrX_-_157568983 | 0.23 |
ENSMUST00000065806.4
|
Yy2
|
Yy2 transcription factor |
| chr1_+_70725902 | 0.23 |
ENSMUST00000161937.1
ENSMUST00000162182.1 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr3_-_144760841 | 0.23 |
ENSMUST00000059091.5
|
Clca1
|
chloride channel calcium activated 1 |
| chr7_+_110768169 | 0.23 |
ENSMUST00000170374.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr1_-_17097839 | 0.23 |
ENSMUST00000038382.4
|
Jph1
|
junctophilin 1 |
| chr11_-_11970540 | 0.23 |
ENSMUST00000109653.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr11_-_69858723 | 0.23 |
ENSMUST00000001626.3
ENSMUST00000108626.1 |
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr13_+_81657732 | 0.23 |
ENSMUST00000049055.6
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr12_-_4592927 | 0.22 |
ENSMUST00000170816.1
|
Gm3625
|
predicted gene 3625 |
| chr2_+_30845059 | 0.22 |
ENSMUST00000041659.5
|
Prrx2
|
paired related homeobox 2 |
| chr15_-_95528228 | 0.22 |
ENSMUST00000075275.2
|
Nell2
|
NEL-like 2 |
| chr16_+_21794320 | 0.22 |
ENSMUST00000181780.1
ENSMUST00000181960.1 |
1300002E11Rik
|
RIKEN cDNA 1300002E11 gene |
| chr10_-_80421847 | 0.22 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr3_-_106024911 | 0.22 |
ENSMUST00000066537.3
ENSMUST00000054973.3 |
Chi3l7
|
chitinase 3-like 7 |
| chr16_+_77014069 | 0.22 |
ENSMUST00000023580.6
|
Usp25
|
ubiquitin specific peptidase 25 |
| chr2_+_130277157 | 0.21 |
ENSMUST00000028890.8
ENSMUST00000159373.1 |
Nop56
|
NOP56 ribonucleoprotein |
| chr14_+_79481164 | 0.21 |
ENSMUST00000040131.5
|
Elf1
|
E74-like factor 1 |
| chr17_-_17855188 | 0.21 |
ENSMUST00000003762.6
|
Has1
|
hyaluronan synthase1 |
| chr11_-_61719946 | 0.21 |
ENSMUST00000151780.1
ENSMUST00000148584.1 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr10_+_127000709 | 0.21 |
ENSMUST00000026500.5
ENSMUST00000142698.1 |
Avil
|
advillin |
| chr5_+_110110068 | 0.21 |
ENSMUST00000112528.1
|
Zfp605
|
zinc finger protein 605 |
| chr3_+_27154020 | 0.21 |
ENSMUST00000181124.1
|
1700125G22Rik
|
RIKEN cDNA 1700125G22 gene |
| chr5_-_21785115 | 0.21 |
ENSMUST00000115193.1
ENSMUST00000115192.1 ENSMUST00000115195.1 ENSMUST00000030771.5 |
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_-_79481268 | 0.21 |
ENSMUST00000022601.5
|
Wbp4
|
WW domain binding protein 4 |
| chrX_+_72918557 | 0.21 |
ENSMUST00000033715.4
|
Nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr2_+_103424056 | 0.21 |
ENSMUST00000028609.7
|
Elf5
|
E74-like factor 5 |
| chr11_+_46454921 | 0.20 |
ENSMUST00000020668.8
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chrX_-_51205990 | 0.20 |
ENSMUST00000114876.2
|
Mbnl3
|
muscleblind-like 3 (Drosophila) |
| chr8_+_47824459 | 0.20 |
ENSMUST00000038693.6
|
Cldn22
|
claudin 22 |
| chr11_-_33513626 | 0.20 |
ENSMUST00000037522.7
|
Ranbp17
|
RAN binding protein 17 |
| chr13_+_76579681 | 0.20 |
ENSMUST00000109589.2
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr6_-_118562226 | 0.20 |
ENSMUST00000112830.1
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr13_-_111490111 | 0.20 |
ENSMUST00000047627.7
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr3_+_79591356 | 0.20 |
ENSMUST00000029382.7
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D) |
| chr2_-_144527341 | 0.20 |
ENSMUST00000163701.1
ENSMUST00000081982.5 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr6_-_66896521 | 0.20 |
ENSMUST00000065878.3
|
4930597O21Rik
|
RIKEN cDNA 4930597O21 gene |
| chr1_+_137966529 | 0.19 |
ENSMUST00000182158.1
|
A430106G13Rik
|
RIKEN cDNA A430106G13 gene |
| chr6_-_97205549 | 0.19 |
ENSMUST00000164744.1
ENSMUST00000089287.5 |
Uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr5_+_17574726 | 0.19 |
ENSMUST00000169603.1
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_+_138963709 | 0.19 |
ENSMUST00000168527.1
|
Dennd1b
|
DENN/MADD domain containing 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0045041 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 1.2 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.3 | 1.7 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 0.7 | GO:0051659 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.8 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 0.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 1.4 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.4 | GO:0033624 | negative regulation of integrin activation(GO:0033624) |
| 0.1 | 1.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.4 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.4 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.6 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.1 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:0033368 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.2 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 0.3 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 0.3 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.4 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.1 | 0.2 | GO:1903660 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 1.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.9 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 1.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.1 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.1 | 0.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.7 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.4 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.7 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0009196 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.2 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.2 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:0071374 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:1903405 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 1.9 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.2 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.0 | GO:1904305 | negative regulation of gastro-intestinal system smooth muscle contraction(GO:1904305) negative regulation of small intestine smooth muscle contraction(GO:1904348) |
| 0.0 | 0.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0005818 | aster(GO:0005818) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.6 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 1.8 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 0.7 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.9 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 1.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.0 | 0.1 | GO:0050145 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.0 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |