Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Insm1
Z-value: 1.48
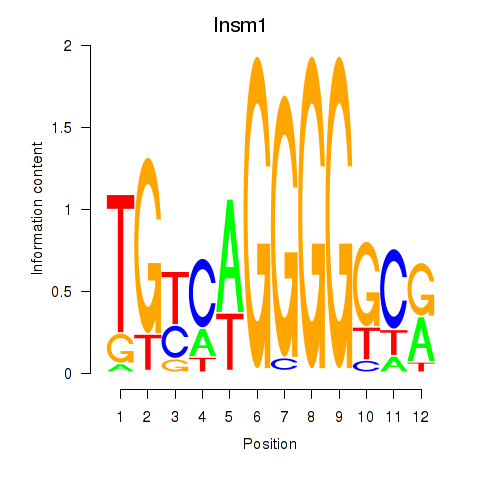
Transcription factors associated with Insm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Insm1
|
ENSMUSG00000068154.4 | insulinoma-associated 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Insm1 | mm10_v2_chr2_+_146221921_146221921 | 0.41 | 1.4e-02 | Click! |
Activity profile of Insm1 motif
Sorted Z-values of Insm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_78914216 | 6.39 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr7_+_78913765 | 6.05 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr7_+_79660196 | 5.21 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr10_-_19851459 | 4.88 |
ENSMUST00000059805.4
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr7_-_126369543 | 4.29 |
ENSMUST00000032997.6
|
Lat
|
linker for activation of T cells |
| chr13_-_21780616 | 4.14 |
ENSMUST00000080511.2
|
Hist1h1b
|
histone cluster 1, H1b |
| chr7_+_27473761 | 3.38 |
ENSMUST00000068641.6
|
Sertad3
|
SERTA domain containing 3 |
| chr4_-_44710408 | 3.26 |
ENSMUST00000134968.2
ENSMUST00000173821.1 ENSMUST00000174319.1 ENSMUST00000173733.1 ENSMUST00000172866.1 ENSMUST00000165417.2 ENSMUST00000107825.2 ENSMUST00000102932.3 ENSMUST00000107827.2 ENSMUST00000107826.2 |
Pax5
|
paired box gene 5 |
| chr8_+_95055094 | 3.15 |
ENSMUST00000058479.6
|
Ccdc135
|
coiled-coil domain containing 135 |
| chr7_+_127746775 | 3.09 |
ENSMUST00000033081.7
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr2_+_109280738 | 3.09 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr4_-_108383349 | 3.06 |
ENSMUST00000053157.6
|
Fam159a
|
family with sequence similarity 159, member A |
| chr12_-_12940600 | 3.02 |
ENSMUST00000130990.1
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr7_-_110982049 | 2.97 |
ENSMUST00000142368.1
|
Mrvi1
|
MRV integration site 1 |
| chr2_+_91082362 | 2.87 |
ENSMUST00000169852.1
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_-_110982443 | 2.85 |
ENSMUST00000005751.6
|
Mrvi1
|
MRV integration site 1 |
| chr6_+_38109320 | 2.72 |
ENSMUST00000031851.3
|
Tmem213
|
transmembrane protein 213 |
| chr7_+_144838590 | 2.65 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chr19_+_4154606 | 2.61 |
ENSMUST00000061086.8
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr4_-_88033328 | 2.58 |
ENSMUST00000078090.5
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr4_+_46450892 | 2.58 |
ENSMUST00000102926.4
|
Anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr17_-_25952565 | 2.58 |
ENSMUST00000162431.1
|
A930017K11Rik
|
RIKEN cDNA A930017K11 gene |
| chr4_-_3938354 | 2.50 |
ENSMUST00000003369.3
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr15_-_75888754 | 2.46 |
ENSMUST00000184858.1
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr12_-_70111920 | 2.43 |
ENSMUST00000169074.1
ENSMUST00000021468.7 |
Nin
|
ninein |
| chr5_-_137741601 | 2.43 |
ENSMUST00000119498.1
ENSMUST00000061789.7 |
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr4_+_134315112 | 2.28 |
ENSMUST00000105875.1
ENSMUST00000030638.6 |
Trim63
|
tripartite motif-containing 63 |
| chr7_+_141131268 | 2.23 |
ENSMUST00000026568.8
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr2_+_29889785 | 2.18 |
ENSMUST00000113763.1
ENSMUST00000113757.1 ENSMUST00000113756.1 ENSMUST00000133233.1 ENSMUST00000113759.2 ENSMUST00000113755.1 ENSMUST00000137558.1 ENSMUST00000046571.7 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr5_-_137741102 | 2.15 |
ENSMUST00000149512.1
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr2_+_29889720 | 2.10 |
ENSMUST00000113767.1
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr2_+_25242929 | 1.92 |
ENSMUST00000114355.1
ENSMUST00000060818.1 |
Rnf208
|
ring finger protein 208 |
| chr10_-_116473875 | 1.88 |
ENSMUST00000068233.4
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr7_-_126463100 | 1.86 |
ENSMUST00000032974.6
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chrX_+_9199865 | 1.82 |
ENSMUST00000069763.2
|
Lancl3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr4_+_119637704 | 1.76 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr6_-_131316398 | 1.76 |
ENSMUST00000121078.1
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr7_+_4740137 | 1.69 |
ENSMUST00000130215.1
ENSMUST00000108582.3 |
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr3_-_104818266 | 1.68 |
ENSMUST00000168015.1
|
Mov10
|
Moloney leukemia virus 10 |
| chr7_-_110982169 | 1.67 |
ENSMUST00000154466.1
|
Mrvi1
|
MRV integration site 1 |
| chr10_-_42276688 | 1.66 |
ENSMUST00000175881.1
ENSMUST00000056974.3 |
Foxo3
|
forkhead box O3 |
| chr3_-_104818539 | 1.66 |
ENSMUST00000106774.1
ENSMUST00000106775.1 ENSMUST00000166979.1 ENSMUST00000136148.1 |
Mov10
|
Moloney leukemia virus 10 |
| chr7_+_4740111 | 1.66 |
ENSMUST00000098853.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr2_-_152831665 | 1.63 |
ENSMUST00000156688.1
ENSMUST00000007803.5 |
Bcl2l1
|
BCL2-like 1 |
| chr7_+_4740178 | 1.58 |
ENSMUST00000108583.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr2_+_11172382 | 1.58 |
ENSMUST00000028118.3
|
Prkcq
|
protein kinase C, theta |
| chr11_+_85171096 | 1.48 |
ENSMUST00000018623.3
|
1700125H20Rik
|
RIKEN cDNA 1700125H20 gene |
| chr6_+_128438757 | 1.48 |
ENSMUST00000144745.1
|
Gm10069
|
predicted gene 10069 |
| chr3_-_104818224 | 1.44 |
ENSMUST00000002297.5
|
Mov10
|
Moloney leukemia virus 10 |
| chr2_-_152831112 | 1.35 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr3_+_134236483 | 1.30 |
ENSMUST00000181904.1
ENSMUST00000053048.9 |
Cxxc4
|
CXXC finger 4 |
| chr6_-_38109548 | 1.29 |
ENSMUST00000114908.1
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr5_-_122900267 | 1.28 |
ENSMUST00000031435.7
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr17_-_35516780 | 1.27 |
ENSMUST00000160885.1
ENSMUST00000159009.1 ENSMUST00000161012.1 |
Tcf19
|
transcription factor 19 |
| chr4_-_153482768 | 1.22 |
ENSMUST00000105646.2
|
Ajap1
|
adherens junction associated protein 1 |
| chrX_+_68678712 | 1.21 |
ENSMUST00000114654.1
ENSMUST00000114655.1 ENSMUST00000114657.2 ENSMUST00000114653.1 |
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr2_+_52072823 | 1.19 |
ENSMUST00000112693.2
ENSMUST00000069794.5 |
Rif1
|
Rap1 interacting factor 1 homolog (yeast) |
| chr1_-_172027269 | 1.16 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chrX_-_166585679 | 1.14 |
ENSMUST00000000412.2
|
Egfl6
|
EGF-like-domain, multiple 6 |
| chrX_+_68678541 | 1.13 |
ENSMUST00000088546.5
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr15_-_99370427 | 1.12 |
ENSMUST00000081224.7
ENSMUST00000120633.1 ENSMUST00000088233.6 |
Fmnl3
|
formin-like 3 |
| chr7_-_19715395 | 1.09 |
ENSMUST00000032555.9
ENSMUST00000093552.5 |
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr8_+_81342556 | 1.08 |
ENSMUST00000172167.1
ENSMUST00000169116.1 ENSMUST00000109852.2 ENSMUST00000172031.1 |
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr4_+_130047840 | 1.05 |
ENSMUST00000044565.8
ENSMUST00000132251.1 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr17_+_3114957 | 1.03 |
ENSMUST00000076734.6
|
Scaf8
|
SR-related CTD-associated factor 8 |
| chr10_+_80805233 | 1.02 |
ENSMUST00000036016.4
|
Amh
|
anti-Mullerian hormone |
| chr7_+_66060338 | 0.98 |
ENSMUST00000153609.1
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr1_+_87620306 | 0.98 |
ENSMUST00000169754.1
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr1_+_87620334 | 0.94 |
ENSMUST00000042275.8
ENSMUST00000168783.1 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chrX_+_153006461 | 0.94 |
ENSMUST00000095755.3
|
Usp51
|
ubiquitin specific protease 51 |
| chr7_+_100372224 | 0.93 |
ENSMUST00000051777.8
ENSMUST00000098259.4 |
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr1_-_172027251 | 0.85 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr14_-_55116935 | 0.84 |
ENSMUST00000022819.5
|
Jph4
|
junctophilin 4 |
| chr7_-_118995211 | 0.81 |
ENSMUST00000008878.8
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr19_+_6975048 | 0.79 |
ENSMUST00000070850.6
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr7_+_5056706 | 0.75 |
ENSMUST00000144802.1
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr6_+_4902913 | 0.73 |
ENSMUST00000175889.1
ENSMUST00000168998.2 |
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A |
| chr4_+_129960760 | 0.72 |
ENSMUST00000139884.1
|
1700003M07Rik
|
RIKEN cDNA 1700003M07 gene |
| chr12_+_77239036 | 0.71 |
ENSMUST00000062804.7
|
Fut8
|
fucosyltransferase 8 |
| chr8_+_71469186 | 0.70 |
ENSMUST00000124745.1
ENSMUST00000138892.1 ENSMUST00000147642.1 |
Dda1
|
DET1 and DDB1 associated 1 |
| chr12_+_3891728 | 0.69 |
ENSMUST00000172689.1
ENSMUST00000111186.1 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr14_-_20668269 | 0.69 |
ENSMUST00000057090.5
ENSMUST00000117386.1 |
Synpo2l
|
synaptopodin 2-like |
| chr7_+_4690760 | 0.68 |
ENSMUST00000048248.7
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr10_+_75518042 | 0.66 |
ENSMUST00000020397.8
|
Snrpd3
|
small nuclear ribonucleoprotein D3 |
| chr3_+_145938004 | 0.61 |
ENSMUST00000039571.7
|
2410004B18Rik
|
RIKEN cDNA 2410004B18 gene |
| chr7_-_45016224 | 0.56 |
ENSMUST00000085383.2
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr18_+_34777008 | 0.51 |
ENSMUST00000043775.7
|
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr4_-_130174691 | 0.46 |
ENSMUST00000132545.2
ENSMUST00000175992.1 ENSMUST00000105999.2 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr9_-_44881274 | 0.44 |
ENSMUST00000002095.3
ENSMUST00000114689.1 ENSMUST00000128768.1 |
Kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr5_+_53590453 | 0.43 |
ENSMUST00000113865.1
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_-_125434048 | 0.42 |
ENSMUST00000169485.1
|
Dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr14_+_54625305 | 0.41 |
ENSMUST00000097177.4
|
Psmb11
|
proteasome (prosome, macropain) subunit, beta type, 11 |
| chr2_-_127521358 | 0.41 |
ENSMUST00000028850.8
ENSMUST00000103215.4 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr7_-_30552255 | 0.41 |
ENSMUST00000108165.1
ENSMUST00000153594.1 |
BC053749
|
cDNA sequence BC053749 |
| chr8_+_124897877 | 0.41 |
ENSMUST00000034467.5
|
Sprtn
|
SprT-like N-terminal domain |
| chr7_+_4690604 | 0.39 |
ENSMUST00000120836.1
|
Brsk1
|
BR serine/threonine kinase 1 |
| chrX_+_68678624 | 0.39 |
ENSMUST00000114656.1
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr17_-_46680870 | 0.38 |
ENSMUST00000165007.1
ENSMUST00000071841.5 |
Klhdc3
|
kelch domain containing 3 |
| chr12_-_114406773 | 0.35 |
ENSMUST00000177949.1
|
Ighv6-4
|
immunoglobulin heavy variable V6-4 |
| chr7_+_100159241 | 0.32 |
ENSMUST00000032967.3
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr15_+_78983041 | 0.32 |
ENSMUST00000109687.1
ENSMUST00000109688.1 ENSMUST00000130663.2 |
Triobp
|
TRIO and F-actin binding protein |
| chr4_-_134245579 | 0.32 |
ENSMUST00000030644.7
|
Zfp593
|
zinc finger protein 593 |
| chr17_-_32166879 | 0.28 |
ENSMUST00000087723.3
|
Notch3
|
notch 3 |
| chr2_-_72980402 | 0.28 |
ENSMUST00000066003.6
ENSMUST00000102689.3 |
Sp3
|
trans-acting transcription factor 3 |
| chr5_+_149184648 | 0.27 |
ENSMUST00000122160.1
ENSMUST00000100410.3 ENSMUST00000119685.1 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr11_-_115813621 | 0.25 |
ENSMUST00000041684.4
ENSMUST00000156812.1 |
Caskin2
|
CASK-interacting protein 2 |
| chr5_+_149184555 | 0.23 |
ENSMUST00000050472.9
|
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr17_+_47688992 | 0.22 |
ENSMUST00000156118.1
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr6_+_42286676 | 0.22 |
ENSMUST00000031894.6
|
Clcn1
|
chloride channel 1 |
| chr12_+_51593315 | 0.19 |
ENSMUST00000164782.2
ENSMUST00000085412.5 |
Coch
|
coagulation factor C homolog (Limulus polyphemus) |
| chr15_+_100423193 | 0.18 |
ENSMUST00000148928.1
|
Gm5475
|
predicted gene 5475 |
| chr6_+_4903350 | 0.17 |
ENSMUST00000175962.1
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A |
| chr8_+_105413614 | 0.17 |
ENSMUST00000109355.2
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr5_+_77265454 | 0.15 |
ENSMUST00000080359.5
|
Rest
|
RE1-silencing transcription factor |
| chr9_+_106477269 | 0.14 |
ENSMUST00000047721.8
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast) |
| chr5_+_149184678 | 0.12 |
ENSMUST00000139474.1
ENSMUST00000117878.1 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr11_+_106084577 | 0.12 |
ENSMUST00000002044.9
|
Map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr6_+_42286709 | 0.11 |
ENSMUST00000163936.1
|
Clcn1
|
chloride channel 1 |
| chr6_+_6863269 | 0.10 |
ENSMUST00000160937.2
ENSMUST00000171311.1 |
Dlx6
|
distal-less homeobox 6 |
| chr7_+_48789003 | 0.10 |
ENSMUST00000118927.1
ENSMUST00000125280.1 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr16_-_67620880 | 0.09 |
ENSMUST00000114292.1
ENSMUST00000120898.1 |
Cadm2
|
cell adhesion molecule 2 |
| chr12_+_95692212 | 0.09 |
ENSMUST00000057324.3
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_192190771 | 0.09 |
ENSMUST00000078470.5
ENSMUST00000110844.1 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr11_-_115699461 | 0.08 |
ENSMUST00000106497.1
|
Grb2
|
growth factor receptor bound protein 2 |
| chr2_+_178141920 | 0.08 |
ENSMUST00000103066.3
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr7_+_6286589 | 0.07 |
ENSMUST00000170776.1
|
Zfp667
|
zinc finger protein 667 |
| chr7_+_6286573 | 0.07 |
ENSMUST00000086327.5
|
Zfp667
|
zinc finger protein 667 |
| chr7_+_5057161 | 0.06 |
ENSMUST00000045543.5
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr2_+_28205648 | 0.05 |
ENSMUST00000102879.3
ENSMUST00000028177.4 |
Olfm1
|
olfactomedin 1 |
| chr10_-_82623190 | 0.03 |
ENSMUST00000183363.1
ENSMUST00000079648.5 ENSMUST00000185168.1 |
1190007I07Rik
|
RIKEN cDNA 1190007I07 gene |
| chr10_-_116473418 | 0.02 |
ENSMUST00000087965.4
ENSMUST00000164271.1 |
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr9_+_108808356 | 0.01 |
ENSMUST00000035218.7
|
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr7_+_141461075 | 0.01 |
ENSMUST00000053670.5
|
Efcab4a
|
EF-hand calcium binding domain 4A |
| chr7_+_26061495 | 0.00 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr2_+_174330006 | 0.00 |
ENSMUST00000109085.1
ENSMUST00000109087.1 ENSMUST00000109084.1 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
Network of associatons between targets according to the STRING database.
First level regulatory network of Insm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.6 | 4.9 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 1.2 | 4.8 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.9 | 2.7 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.8 | 2.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 3.0 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.7 | 2.0 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.7 | 5.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.6 | 1.9 | GO:0090076 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.6 | 2.9 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.6 | 2.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.5 | 7.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.5 | 1.9 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.5 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.3 | 1.0 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 | 2.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.3 | 2.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 1.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 3.0 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 4.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.7 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.2 | 0.9 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 0.4 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) regulation of cell adhesion involved in heart morphogenesis(GO:0061344) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 0.2 | 3.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.2 | 0.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 0.7 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 1.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 2.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.6 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 2.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 3.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 2.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.4 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.5 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.8 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 1.8 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.3 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 1.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 3.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 3.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.1 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 3.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:1902737 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.5 | 2.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.5 | 2.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 3.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 1.9 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 4.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 13.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 3.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 1.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 5.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 6.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 3.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 4.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.7 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 6.7 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.4 | 2.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 1.8 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.3 | 1.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.3 | 1.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 3.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 2.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 1.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 2.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.2 | 3.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 3.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 1.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 4.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 1.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 4.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 2.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 4.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 12.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 5.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 4.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |