Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Irx5
Z-value: 1.01
Transcription factors associated with Irx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx5
|
ENSMUSG00000031737.10 | Iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx5 | mm10_v2_chr8_+_92357787_92357801 | -0.44 | 7.2e-03 | Click! |
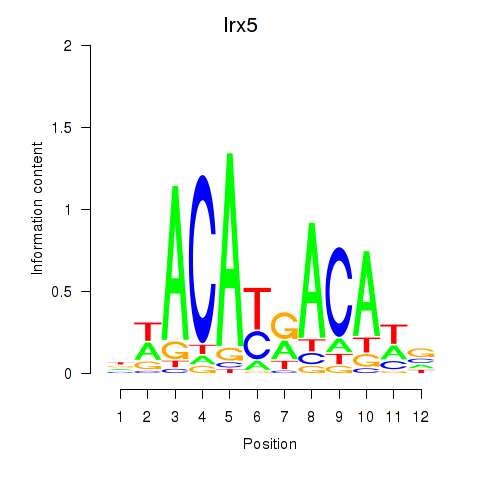
Activity profile of Irx5 motif
Sorted Z-values of Irx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87254804 | 5.53 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr5_-_86926521 | 5.33 |
ENSMUST00000031183.2
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr5_-_87337165 | 5.12 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr7_-_14438538 | 3.97 |
ENSMUST00000168252.2
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr19_-_7802578 | 3.83 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr4_+_43632185 | 3.46 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr4_+_43631935 | 3.00 |
ENSMUST00000030191.8
|
Npr2
|
natriuretic peptide receptor 2 |
| chr15_-_82394022 | 2.13 |
ENSMUST00000170255.1
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr3_+_19957037 | 1.92 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr2_+_173153048 | 1.90 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr19_+_26623419 | 1.88 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_+_20549780 | 1.83 |
ENSMUST00000023832.6
|
Rgn
|
regucalcin |
| chr4_+_104766334 | 1.61 |
ENSMUST00000065072.6
|
C8b
|
complement component 8, beta polypeptide |
| chr4_+_104766308 | 1.60 |
ENSMUST00000031663.3
|
C8b
|
complement component 8, beta polypeptide |
| chr14_+_48120841 | 1.55 |
ENSMUST00000073150.4
|
Peli2
|
pellino 2 |
| chrX_+_38772671 | 1.53 |
ENSMUST00000050744.5
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr12_-_99393010 | 1.51 |
ENSMUST00000177451.1
|
Foxn3
|
forkhead box N3 |
| chr1_+_13668739 | 1.45 |
ENSMUST00000088542.3
|
Xkr9
|
X Kell blood group precursor related family member 9 homolog |
| chr2_-_160872552 | 1.41 |
ENSMUST00000103111.2
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr3_+_19957088 | 1.40 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr12_+_85288591 | 1.34 |
ENSMUST00000059341.4
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr3_+_63963631 | 1.19 |
ENSMUST00000181653.1
|
Gm26850
|
predicted gene, 26850 |
| chr3_+_19957240 | 1.18 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr5_+_23787691 | 1.05 |
ENSMUST00000030852.6
ENSMUST00000120869.1 ENSMUST00000117783.1 ENSMUST00000115113.2 |
Rint1
|
RAD50 interactor 1 |
| chr2_+_104027721 | 1.02 |
ENSMUST00000028603.3
|
Fbxo3
|
F-box protein 3 |
| chr13_+_24943144 | 0.98 |
ENSMUST00000021773.5
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr3_-_88762244 | 0.97 |
ENSMUST00000183267.1
|
Syt11
|
synaptotagmin XI |
| chr2_-_101621033 | 0.93 |
ENSMUST00000090513.4
|
B230118H07Rik
|
RIKEN cDNA B230118H07 gene |
| chr12_-_85288419 | 0.91 |
ENSMUST00000121930.1
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr5_+_138255608 | 0.88 |
ENSMUST00000062067.6
|
Lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr6_+_50110837 | 0.85 |
ENSMUST00000167628.1
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr12_+_55598917 | 0.80 |
ENSMUST00000051857.3
|
Insm2
|
insulinoma-associated 2 |
| chr5_-_88675190 | 0.78 |
ENSMUST00000133532.1
ENSMUST00000150438.1 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr1_-_169969143 | 0.77 |
ENSMUST00000027989.6
ENSMUST00000111353.3 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr16_-_22811399 | 0.75 |
ENSMUST00000040592.4
|
Crygs
|
crystallin, gamma S |
| chr14_+_48446340 | 0.74 |
ENSMUST00000111735.2
|
Tmem260
|
transmembrane protein 260 |
| chr11_-_46389454 | 0.72 |
ENSMUST00000101306.3
|
Itk
|
IL2 inducible T cell kinase |
| chr1_-_132707304 | 0.72 |
ENSMUST00000043189.7
|
Nfasc
|
neurofascin |
| chr4_-_43653542 | 0.66 |
ENSMUST00000084646.4
|
Spag8
|
sperm associated antigen 8 |
| chr11_-_69395333 | 0.65 |
ENSMUST00000108660.1
ENSMUST00000051620.4 |
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr18_+_76944384 | 0.62 |
ENSMUST00000156454.1
ENSMUST00000150990.1 ENSMUST00000026485.7 ENSMUST00000148955.1 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr14_+_14820765 | 0.62 |
ENSMUST00000112631.2
ENSMUST00000178538.1 ENSMUST00000112630.2 |
Nek10
|
NIMA (never in mitosis gene a)- related kinase 10 |
| chr4_-_89311021 | 0.61 |
ENSMUST00000097981.4
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr7_-_103840427 | 0.60 |
ENSMUST00000106866.1
|
Hbb-bh2
|
hemoglobin beta, bh2 |
| chr4_-_43653560 | 0.59 |
ENSMUST00000107870.2
|
Spag8
|
sperm associated antigen 8 |
| chr11_-_28339867 | 0.58 |
ENSMUST00000042595.6
|
Gm6685
|
predicted pseudogene 6685 |
| chr11_-_121519326 | 0.58 |
ENSMUST00000092298.5
|
Zfp750
|
zinc finger protein 750 |
| chr5_-_100429503 | 0.56 |
ENSMUST00000181873.1
ENSMUST00000180779.1 |
5430416N02Rik
|
RIKEN cDNA 5430416N02 gene |
| chr16_-_55838827 | 0.56 |
ENSMUST00000096026.2
ENSMUST00000036273.6 ENSMUST00000114457.1 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr3_-_79737794 | 0.55 |
ENSMUST00000078527.6
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_+_104027823 | 0.53 |
ENSMUST00000111135.1
ENSMUST00000111136.1 ENSMUST00000102565.3 |
Fbxo3
|
F-box protein 3 |
| chr12_-_108893197 | 0.49 |
ENSMUST00000161154.1
ENSMUST00000161410.1 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr16_-_19408078 | 0.48 |
ENSMUST00000052516.4
|
Olfr165
|
olfactory receptor 165 |
| chr1_+_58278309 | 0.48 |
ENSMUST00000114366.1
|
Aox2
|
aldehyde oxidase 2 |
| chr12_-_67222549 | 0.47 |
ENSMUST00000037181.8
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr5_+_87666200 | 0.44 |
ENSMUST00000094641.4
|
Csn1s1
|
casein alpha s1 |
| chrX_+_134187492 | 0.44 |
ENSMUST00000064476.4
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr11_+_78176711 | 0.41 |
ENSMUST00000098545.5
|
Tlcd1
|
TLC domain containing 1 |
| chr16_+_30065333 | 0.39 |
ENSMUST00000023171.7
|
Hes1
|
hairy and enhancer of split 1 (Drosophila) |
| chr18_-_35215008 | 0.39 |
ENSMUST00000091636.3
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr11_+_69395647 | 0.38 |
ENSMUST00000144531.1
|
Lsmd1
|
LSM domain containing 1 |
| chr4_-_43700807 | 0.38 |
ENSMUST00000055545.3
|
Olfr70
|
olfactory receptor 70 |
| chr5_+_139543889 | 0.36 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr6_+_121183667 | 0.36 |
ENSMUST00000118234.1
ENSMUST00000088561.3 ENSMUST00000137432.1 ENSMUST00000120066.1 |
Pex26
|
peroxisomal biogenesis factor 26 |
| chr13_-_106547060 | 0.36 |
ENSMUST00000076359.2
|
Dph3b-ps
|
DPH3B, KTI11 homolog B (S. cerevisiae), pseudogene |
| chr17_-_35643684 | 0.36 |
ENSMUST00000095467.3
|
Dpcr1
|
diffuse panbronchiolitis critical region 1 (human) |
| chr3_+_87357874 | 0.33 |
ENSMUST00000015998.6
|
Cd5l
|
CD5 antigen-like |
| chr12_+_69893105 | 0.32 |
ENSMUST00000021466.8
|
Atl1
|
atlastin GTPase 1 |
| chr12_+_71048338 | 0.31 |
ENSMUST00000135709.1
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_+_87025550 | 0.31 |
ENSMUST00000024959.3
ENSMUST00000160269.1 |
Cript
|
cysteine-rich PDZ-binding protein |
| chr17_+_21657582 | 0.26 |
ENSMUST00000039726.7
|
3110052M02Rik
|
RIKEN cDNA 3110052M02 gene |
| chr4_-_132144486 | 0.26 |
ENSMUST00000056336.1
|
Oprd1
|
opioid receptor, delta 1 |
| chr7_+_56239743 | 0.25 |
ENSMUST00000032633.5
ENSMUST00000156886.1 |
Oca2
|
oculocutaneous albinism II |
| chr11_-_99285260 | 0.25 |
ENSMUST00000017255.3
|
Krt24
|
keratin 24 |
| chr14_-_14120904 | 0.23 |
ENSMUST00000022256.3
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr7_+_102803480 | 0.23 |
ENSMUST00000061096.6
|
Olfr564
|
olfactory receptor 564 |
| chr14_-_59142886 | 0.21 |
ENSMUST00000022548.3
ENSMUST00000162674.1 ENSMUST00000159858.1 ENSMUST00000162271.1 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr3_+_139205887 | 0.19 |
ENSMUST00000062306.6
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr9_+_78051938 | 0.18 |
ENSMUST00000024104.7
|
Gcm1
|
glial cells missing homolog 1 (Drosophila) |
| chr9_+_32393963 | 0.17 |
ENSMUST00000172015.1
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr8_+_4240105 | 0.16 |
ENSMUST00000110994.1
ENSMUST00000110995.1 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr15_+_102990576 | 0.13 |
ENSMUST00000001703.6
|
Hoxc8
|
homeobox C8 |
| chr17_-_22007301 | 0.13 |
ENSMUST00000075018.3
|
Gm9772
|
predicted gene 9772 |
| chr9_-_7835255 | 0.12 |
ENSMUST00000074246.6
|
Birc2
|
baculoviral IAP repeat-containing 2 |
| chr10_-_130112788 | 0.11 |
ENSMUST00000081469.1
|
Olfr823
|
olfactory receptor 823 |
| chr16_-_56024628 | 0.10 |
ENSMUST00000119981.1
ENSMUST00000096021.3 |
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr6_+_40442863 | 0.10 |
ENSMUST00000038907.8
ENSMUST00000141490.1 |
Wee2
|
WEE1 homolog 2 (S. pombe) |
| chr8_+_85171322 | 0.09 |
ENSMUST00000076896.5
|
Gm6531
|
predicted gene 6531 |
| chr5_-_144026596 | 0.08 |
ENSMUST00000031622.6
ENSMUST00000110702.1 |
Ocm
|
oncomodulin |
| chr9_-_110624361 | 0.08 |
ENSMUST00000035069.9
|
Nradd
|
neurotrophin receptor associated death domain |
| chr7_+_34095431 | 0.05 |
ENSMUST00000108083.1
ENSMUST00000174820.1 |
Scgb1b30
|
secretoglobin, family 1B, member 30 |
| chr2_-_79908428 | 0.05 |
ENSMUST00000102652.3
ENSMUST00000102651.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_173982842 | 0.04 |
ENSMUST00000000266.7
|
Ifi202b
|
interferon activated gene 202B |
| chr3_+_92352141 | 0.04 |
ENSMUST00000068399.1
|
Sprr2e
|
small proline-rich protein 2E |
| chr13_+_94173992 | 0.03 |
ENSMUST00000121618.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr7_+_45785331 | 0.02 |
ENSMUST00000120005.1
ENSMUST00000123585.1 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr13_-_64497792 | 0.02 |
ENSMUST00000180282.1
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.7 | 6.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.6 | 1.9 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.5 | 1.8 | GO:0051344 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) positive regulation of calcium-transporting ATPase activity(GO:1901896) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.3 | 1.0 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.3 | 5.1 | GO:0006063 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.3 | 1.0 | GO:0006507 | GPI anchor release(GO:0006507) |
| 0.3 | 1.0 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 3.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 3.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.6 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 0.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 4.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.4 | GO:0042668 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 1.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.4 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.6 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein K48-linked ubiquitination(GO:1902524) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 1.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:1901979 | renal sodium ion absorption(GO:0070294) regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.0 | 1.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.8 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.3 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.3 | 3.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 5.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 0.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 4.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 5.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 4.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 16.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 3.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 1.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.5 | GO:0016623 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 1.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 4.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 3.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |