Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Isl1
Z-value: 0.43
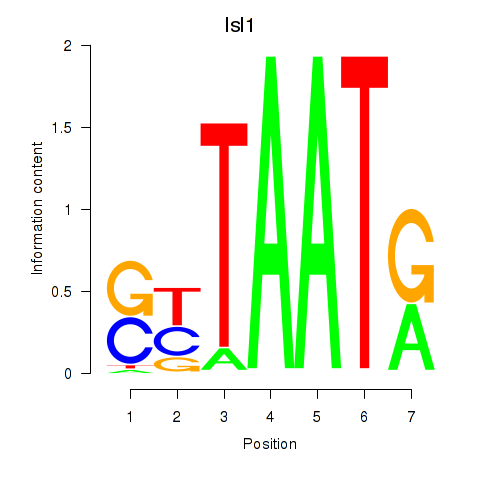
Transcription factors associated with Isl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl1
|
ENSMUSG00000042258.7 | ISL1 transcription factor, LIM/homeodomain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | mm10_v2_chr13_-_116309639_116309699 | 0.08 | 6.4e-01 | Click! |
Activity profile of Isl1 motif
Sorted Z-values of Isl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_105568298 | 0.98 |
ENSMUST00000005849.5
|
Agrp
|
agouti related protein |
| chrX_+_164090187 | 0.93 |
ENSMUST00000015545.3
|
Tmem27
|
transmembrane protein 27 |
| chr10_-_19014549 | 0.59 |
ENSMUST00000146388.1
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_+_69963452 | 0.56 |
ENSMUST00000110560.1
|
Gm3086
|
predicted gene 3086 |
| chr6_+_142298419 | 0.51 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr1_-_79440039 | 0.48 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chr13_+_76579670 | 0.45 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr18_-_68429235 | 0.44 |
ENSMUST00000052347.6
|
Mc2r
|
melanocortin 2 receptor |
| chr9_-_95815389 | 0.43 |
ENSMUST00000119760.1
|
Pls1
|
plastin 1 (I-isoform) |
| chr1_-_75133866 | 0.43 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr3_-_75270073 | 0.41 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr8_-_122460666 | 0.41 |
ENSMUST00000006762.5
|
Snai3
|
snail homolog 3 (Drosophila) |
| chr17_-_78684262 | 0.41 |
ENSMUST00000145480.1
|
Strn
|
striatin, calmodulin binding protein |
| chr14_+_124005355 | 0.41 |
ENSMUST00000166105.1
|
Gm17615
|
predicted gene, 17615 |
| chr3_+_109573907 | 0.40 |
ENSMUST00000106576.2
|
Vav3
|
vav 3 oncogene |
| chr3_+_121531653 | 0.40 |
ENSMUST00000181070.1
|
A530020G20Rik
|
RIKEN cDNA A530020G20 gene |
| chr16_-_22161450 | 0.37 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr13_-_23991158 | 0.36 |
ENSMUST00000021770.7
|
Scgn
|
secretagogin, EF-hand calcium binding protein |
| chr3_+_10088173 | 0.33 |
ENSMUST00000061419.7
|
Gm9833
|
predicted gene 9833 |
| chr3_-_7613427 | 0.33 |
ENSMUST00000168269.2
|
Il7
|
interleukin 7 |
| chr1_-_89933290 | 0.31 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr17_-_51810866 | 0.31 |
ENSMUST00000176669.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr9_+_3404058 | 0.31 |
ENSMUST00000027027.5
|
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr14_-_70630149 | 0.29 |
ENSMUST00000022694.9
|
Dmtn
|
dematin actin binding protein |
| chr4_-_82850721 | 0.28 |
ENSMUST00000139401.1
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr2_+_125152505 | 0.28 |
ENSMUST00000110494.2
ENSMUST00000028630.2 ENSMUST00000110495.2 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr15_+_103018936 | 0.26 |
ENSMUST00000165375.1
|
Hoxc4
|
homeobox C4 |
| chr7_-_28547109 | 0.25 |
ENSMUST00000057974.3
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chrX_+_96455359 | 0.25 |
ENSMUST00000033553.7
|
Heph
|
hephaestin |
| chr3_+_84666192 | 0.25 |
ENSMUST00000107682.1
|
Tmem154
|
transmembrane protein 154 |
| chr14_-_62761112 | 0.24 |
ENSMUST00000053959.6
|
Ints6
|
integrator complex subunit 6 |
| chr6_-_24956106 | 0.23 |
ENSMUST00000127247.2
|
Tmem229a
|
transmembrane protein 229A |
| chr18_-_47333311 | 0.23 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr11_+_68692070 | 0.23 |
ENSMUST00000108673.1
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr9_+_53771499 | 0.22 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr13_-_103764502 | 0.22 |
ENSMUST00000074616.5
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr10_-_35711891 | 0.21 |
ENSMUST00000080898.2
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chrX_+_164140447 | 0.21 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr4_-_42084291 | 0.21 |
ENSMUST00000177937.1
|
Gm21968
|
predicted gene, 21968 |
| chr8_-_26119125 | 0.21 |
ENSMUST00000037182.7
|
Hook3
|
hook homolog 3 (Drosophila) |
| chrX_+_114474312 | 0.21 |
ENSMUST00000113371.1
ENSMUST00000040504.5 |
Klhl4
|
kelch-like 4 |
| chr17_-_24533709 | 0.20 |
ENSMUST00000061764.7
|
Rab26
|
RAB26, member RAS oncogene family |
| chr19_-_53371766 | 0.20 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr18_+_69925542 | 0.20 |
ENSMUST00000080050.5
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr4_-_41045381 | 0.19 |
ENSMUST00000054945.7
|
Aqp7
|
aquaporin 7 |
| chr2_-_58160495 | 0.18 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr15_-_34356421 | 0.17 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr1_+_139422196 | 0.17 |
ENSMUST00000039867.7
|
Zbtb41
|
zinc finger and BTB domain containing 41 homolog |
| chrX_+_106920618 | 0.17 |
ENSMUST00000060576.7
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr2_-_72986716 | 0.17 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr12_-_27342696 | 0.17 |
ENSMUST00000079063.5
|
Sox11
|
SRY-box containing gene 11 |
| chr3_-_127225917 | 0.16 |
ENSMUST00000182064.1
ENSMUST00000182662.1 |
Ank2
|
ankyrin 2, brain |
| chr9_+_35423582 | 0.16 |
ENSMUST00000154652.1
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr3_+_116594959 | 0.16 |
ENSMUST00000029571.8
|
Sass6
|
spindle assembly 6 homolog (C. elegans) |
| chr11_-_76509419 | 0.16 |
ENSMUST00000094012.4
|
Abr
|
active BCR-related gene |
| chr9_-_112217261 | 0.15 |
ENSMUST00000159451.1
ENSMUST00000162796.1 ENSMUST00000161097.1 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr3_-_127225847 | 0.15 |
ENSMUST00000182726.1
ENSMUST00000182779.1 |
Ank2
|
ankyrin 2, brain |
| chr13_+_93308006 | 0.15 |
ENSMUST00000079086.6
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr9_-_40346290 | 0.15 |
ENSMUST00000121357.1
|
Gramd1b
|
GRAM domain containing 1B |
| chr11_-_86357570 | 0.15 |
ENSMUST00000043624.8
|
Med13
|
mediator complex subunit 13 |
| chr8_+_84723003 | 0.15 |
ENSMUST00000098571.4
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr17_-_45733843 | 0.15 |
ENSMUST00000178179.1
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr5_+_136057267 | 0.13 |
ENSMUST00000006303.4
ENSMUST00000156530.1 |
Upk3bl
|
uroplakin 3B-like |
| chr6_+_30568367 | 0.13 |
ENSMUST00000049251.5
|
Cpa4
|
carboxypeptidase A4 |
| chr13_+_76579681 | 0.12 |
ENSMUST00000109589.2
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr4_+_108719649 | 0.12 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr5_-_5266038 | 0.12 |
ENSMUST00000115451.1
ENSMUST00000115452.1 ENSMUST00000131392.1 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_-_6230127 | 0.12 |
ENSMUST00000004505.2
|
Npc1l1
|
NPC1-like 1 |
| chr13_-_75943812 | 0.12 |
ENSMUST00000022078.5
ENSMUST00000109606.1 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr14_+_52844465 | 0.12 |
ENSMUST00000181360.1
ENSMUST00000183652.1 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr14_+_53640701 | 0.11 |
ENSMUST00000179267.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr15_-_50889691 | 0.11 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr1_-_183147461 | 0.11 |
ENSMUST00000171366.1
|
Disp1
|
dispatched homolog 1 (Drosophila) |
| chr4_-_137118135 | 0.11 |
ENSMUST00000154285.1
|
Gm13001
|
predicted gene 13001 |
| chr16_+_36693972 | 0.11 |
ENSMUST00000023617.6
ENSMUST00000089618.3 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr3_-_104818539 | 0.11 |
ENSMUST00000106774.1
ENSMUST00000106775.1 ENSMUST00000166979.1 ENSMUST00000136148.1 |
Mov10
|
Moloney leukemia virus 10 |
| chr9_+_113812547 | 0.10 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr19_-_8218832 | 0.10 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr10_-_70655934 | 0.10 |
ENSMUST00000162144.1
ENSMUST00000162793.1 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr18_-_6490808 | 0.10 |
ENSMUST00000028100.6
ENSMUST00000050542.5 |
Epc1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr8_-_33747724 | 0.10 |
ENSMUST00000179364.1
|
Smim18
|
small integral membrane protein 18 |
| chr10_+_69534208 | 0.10 |
ENSMUST00000182439.1
ENSMUST00000092434.5 ENSMUST00000092432.5 ENSMUST00000092431.5 ENSMUST00000054167.8 ENSMUST00000047061.6 |
Ank3
|
ankyrin 3, epithelial |
| chr13_+_56703504 | 0.10 |
ENSMUST00000109874.1
|
Smad5
|
SMAD family member 5 |
| chr16_+_36694024 | 0.10 |
ENSMUST00000119464.1
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr4_-_14621805 | 0.09 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr2_+_162943474 | 0.09 |
ENSMUST00000035751.5
ENSMUST00000156954.1 |
L3mbtl1
|
l(3)mbt-like (Drosophila) |
| chr11_-_69822144 | 0.09 |
ENSMUST00000045771.6
|
Spem1
|
sperm maturation 1 |
| chr18_-_15718046 | 0.08 |
ENSMUST00000053017.6
|
Chst9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr3_-_25212720 | 0.08 |
ENSMUST00000091289.3
|
Gm10259
|
predicted pseudogene 10259 |
| chrX_-_136780141 | 0.08 |
ENSMUST00000018739.4
|
Glra4
|
glycine receptor, alpha 4 subunit |
| chr2_+_120476911 | 0.08 |
ENSMUST00000110716.1
ENSMUST00000028748.6 ENSMUST00000090028.5 ENSMUST00000110719.2 |
Capn3
|
calpain 3 |
| chrX_-_23285532 | 0.08 |
ENSMUST00000115319.2
|
Klhl13
|
kelch-like 13 |
| chr3_-_150073620 | 0.08 |
ENSMUST00000057740.5
|
Rpsa-ps10
|
ribosomal protein SA, pseudogene 10 |
| chr15_-_93519499 | 0.08 |
ENSMUST00000109255.2
|
Prickle1
|
prickle homolog 1 (Drosophila) |
| chr10_-_13388753 | 0.08 |
ENSMUST00000105546.1
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr14_-_78536762 | 0.07 |
ENSMUST00000123853.1
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chrX_+_153237748 | 0.07 |
ENSMUST00000112574.2
|
Klf8
|
Kruppel-like factor 8 |
| chrX_+_56779437 | 0.07 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr2_+_158794807 | 0.07 |
ENSMUST00000029186.7
ENSMUST00000109478.2 ENSMUST00000156893.1 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr10_+_26772477 | 0.07 |
ENSMUST00000039557.7
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr8_-_61902669 | 0.07 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr9_-_44767792 | 0.07 |
ENSMUST00000034607.9
|
Arcn1
|
archain 1 |
| chr2_-_71367749 | 0.07 |
ENSMUST00000151937.1
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr6_-_129451906 | 0.07 |
ENSMUST00000037481.7
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr4_-_137766474 | 0.07 |
ENSMUST00000139951.1
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chrX_-_153037549 | 0.07 |
ENSMUST00000051484.3
|
Mageh1
|
melanoma antigen, family H, 1 |
| chr16_-_64786321 | 0.06 |
ENSMUST00000052588.4
|
Zfp654
|
zinc finger protein 654 |
| chr19_-_8839181 | 0.06 |
ENSMUST00000096259.4
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr11_+_68692097 | 0.06 |
ENSMUST00000018887.8
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr4_+_3940747 | 0.06 |
ENSMUST00000119403.1
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_+_103832562 | 0.06 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr4_+_94739276 | 0.06 |
ENSMUST00000073939.6
ENSMUST00000102798.1 |
Tek
|
endothelial-specific receptor tyrosine kinase |
| chr11_-_4594750 | 0.06 |
ENSMUST00000109943.3
|
Mtmr3
|
myotubularin related protein 3 |
| chr2_-_72813665 | 0.06 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr4_-_120951664 | 0.06 |
ENSMUST00000106280.1
|
Zfp69
|
zinc finger protein 69 |
| chr2_+_71211941 | 0.06 |
ENSMUST00000112144.2
ENSMUST00000100028.3 ENSMUST00000112136.1 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr19_+_55741884 | 0.06 |
ENSMUST00000111658.3
ENSMUST00000111654.1 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr12_-_72408934 | 0.06 |
ENSMUST00000078505.7
|
Rtn1
|
reticulon 1 |
| chr10_+_40349265 | 0.05 |
ENSMUST00000044672.4
ENSMUST00000095743.2 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr11_-_99374895 | 0.05 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr10_-_44458715 | 0.04 |
ENSMUST00000039174.4
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr3_+_89520152 | 0.04 |
ENSMUST00000000811.7
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr4_-_94650092 | 0.04 |
ENSMUST00000107101.1
|
Lrrc19
|
leucine rich repeat containing 19 |
| chrX_+_71981522 | 0.04 |
ENSMUST00000114569.1
|
Fate1
|
fetal and adult testis expressed 1 |
| chrX_+_99975570 | 0.04 |
ENSMUST00000113779.1
ENSMUST00000113776.1 ENSMUST00000113775.1 ENSMUST00000113780.1 ENSMUST00000113778.1 ENSMUST00000113781.1 ENSMUST00000113783.1 ENSMUST00000071453.2 ENSMUST00000113777.1 |
Eda
|
ectodysplasin-A |
| chr7_-_118116128 | 0.04 |
ENSMUST00000128482.1
ENSMUST00000131840.1 |
Rps15a
|
ribosomal protein S15A |
| chr5_+_121777929 | 0.04 |
ENSMUST00000160821.1
|
Atxn2
|
ataxin 2 |
| chr15_+_44196135 | 0.04 |
ENSMUST00000038856.6
ENSMUST00000110289.3 |
Trhr
|
thyrotropin releasing hormone receptor |
| chr5_-_107875035 | 0.04 |
ENSMUST00000138111.1
ENSMUST00000112642.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr12_-_12941827 | 0.04 |
ENSMUST00000043396.7
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr15_-_50882806 | 0.04 |
ENSMUST00000184885.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr2_-_152398046 | 0.04 |
ENSMUST00000063332.8
ENSMUST00000182625.1 |
Sox12
|
SRY-box containing gene 12 |
| chr7_-_118116171 | 0.04 |
ENSMUST00000131374.1
|
Rps15a
|
ribosomal protein S15A |
| chr4_+_74013442 | 0.03 |
ENSMUST00000098006.2
ENSMUST00000084474.5 |
Frmd3
|
FERM domain containing 3 |
| chr1_+_87803638 | 0.03 |
ENSMUST00000077772.5
ENSMUST00000177757.1 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chr6_+_139843648 | 0.03 |
ENSMUST00000087657.6
|
Pik3c2g
|
phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide |
| chr12_-_113422730 | 0.03 |
ENSMUST00000177715.1
ENSMUST00000103426.1 |
Ighm
|
immunoglobulin heavy constant mu |
| chr5_-_93045022 | 0.03 |
ENSMUST00000061328.5
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr19_-_37176055 | 0.03 |
ENSMUST00000142973.1
ENSMUST00000154376.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_-_57970067 | 0.03 |
ENSMUST00000164963.1
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr1_-_57970104 | 0.03 |
ENSMUST00000114410.3
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr11_-_60913775 | 0.02 |
ENSMUST00000019075.3
|
Gm16515
|
predicted gene, Gm16515 |
| chr8_+_83666827 | 0.02 |
ENSMUST00000019608.5
|
Ptger1
|
prostaglandin E receptor 1 (subtype EP1) |
| chr19_+_55741810 | 0.02 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr15_+_102990576 | 0.02 |
ENSMUST00000001703.6
|
Hoxc8
|
homeobox C8 |
| chr10_+_18469958 | 0.02 |
ENSMUST00000162891.1
ENSMUST00000100054.3 |
Nhsl1
|
NHS-like 1 |
| chr19_+_55742056 | 0.02 |
ENSMUST00000111659.2
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr9_-_112187898 | 0.02 |
ENSMUST00000178410.1
ENSMUST00000172380.3 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr17_+_27856443 | 0.01 |
ENSMUST00000114849.1
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr2_-_127788854 | 0.01 |
ENSMUST00000028857.7
ENSMUST00000110357.1 |
Nphp1
|
nephronophthisis 1 (juvenile) homolog (human) |
| chr1_-_162898484 | 0.01 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr16_-_97170707 | 0.01 |
ENSMUST00000056102.7
|
Dscam
|
Down syndrome cell adhesion molecule |
| chr18_+_4375583 | 0.01 |
ENSMUST00000025077.6
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr4_-_126325641 | 0.01 |
ENSMUST00000131113.1
|
Tekt2
|
tektin 2 |
| chr14_-_52237572 | 0.01 |
ENSMUST00000089752.4
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr1_+_12718496 | 0.01 |
ENSMUST00000088585.3
|
Sulf1
|
sulfatase 1 |
| chr8_+_25720054 | 0.01 |
ENSMUST00000068916.8
ENSMUST00000139836.1 |
Ppapdc1b
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chrX_+_169879596 | 0.01 |
ENSMUST00000112105.1
ENSMUST00000078947.5 |
Mid1
|
midline 1 |
| chr8_-_54718664 | 0.01 |
ENSMUST00000144711.2
ENSMUST00000093510.2 |
Wdr17
|
WD repeat domain 17 |
| chr4_-_58009117 | 0.00 |
ENSMUST00000102897.3
|
Txndc8
|
thioredoxin domain containing 8 |
| chr5_-_37824580 | 0.00 |
ENSMUST00000063116.9
|
Msx1
|
msh homeobox 1 |
| chr17_-_35046539 | 0.00 |
ENSMUST00000007250.7
|
Msh5
|
mutS homolog 5 (E. coli) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.9 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.2 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.0 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:0070315 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.8 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |