Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
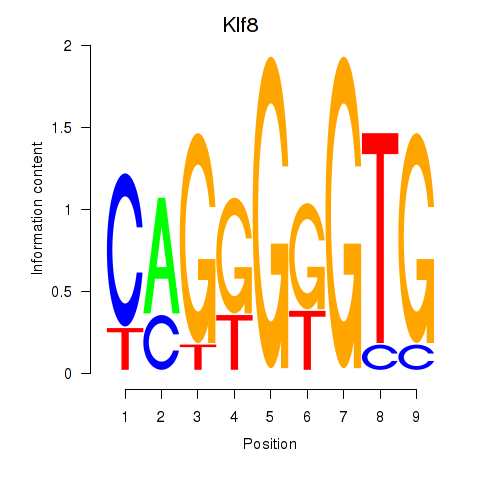
Results for Klf8
Z-value: 0.54
Transcription factors associated with Klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf8
|
ENSMUSG00000041649.7 | Kruppel-like factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf8 | mm10_v2_chrX_+_153237748_153237748 | -0.25 | 1.3e-01 | Click! |
Activity profile of Klf8 motif
Sorted Z-values of Klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_139582790 | 1.05 |
ENSMUST00000106095.2
|
Nkx6-2
|
NK6 homeobox 2 |
| chr15_+_102966794 | 0.94 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr7_-_30924169 | 0.94 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr17_-_70851189 | 0.91 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr3_+_94377432 | 0.83 |
ENSMUST00000107292.1
|
Rorc
|
RAR-related orphan receptor gamma |
| chr11_+_83746940 | 0.77 |
ENSMUST00000070832.2
|
1100001G20Rik
|
RIKEN cDNA 1100001G20 gene |
| chr2_-_168741898 | 0.74 |
ENSMUST00000109176.1
ENSMUST00000178504.1 |
Atp9a
|
ATPase, class II, type 9A |
| chr11_-_102296618 | 0.73 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr3_+_94377505 | 0.72 |
ENSMUST00000098877.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr15_-_89373810 | 0.67 |
ENSMUST00000167643.2
|
Sco2
|
SCO cytochrome oxidase deficient homolog 2 (yeast) |
| chr9_-_110742577 | 0.65 |
ENSMUST00000006005.7
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr1_-_74885322 | 0.63 |
ENSMUST00000159232.1
ENSMUST00000068631.3 |
Fev
|
FEV (ETS oncogene family) |
| chr12_+_84009481 | 0.62 |
ENSMUST00000168120.1
|
Acot1
|
acyl-CoA thioesterase 1 |
| chr7_+_126847908 | 0.62 |
ENSMUST00000147257.1
ENSMUST00000139174.1 |
Doc2a
|
double C2, alpha |
| chr4_+_141368116 | 0.60 |
ENSMUST00000006380.4
|
Fam131c
|
family with sequence similarity 131, member C |
| chr19_-_6067785 | 0.58 |
ENSMUST00000162575.1
ENSMUST00000159084.1 ENSMUST00000161718.1 ENSMUST00000162810.1 ENSMUST00000025713.5 ENSMUST00000113543.2 ENSMUST00000161528.1 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr7_-_127026479 | 0.58 |
ENSMUST00000032916.4
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_+_19004047 | 0.57 |
ENSMUST00000053713.3
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr7_-_19822698 | 0.57 |
ENSMUST00000120537.1
|
Bcl3
|
B cell leukemia/lymphoma 3 |
| chr14_-_30353468 | 0.57 |
ENSMUST00000112249.1
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr6_+_85187438 | 0.57 |
ENSMUST00000045942.8
|
Emx1
|
empty spiracles homeobox 1 |
| chr7_-_31055594 | 0.56 |
ENSMUST00000039909.6
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr10_-_127351753 | 0.54 |
ENSMUST00000059718.4
|
Inhbe
|
inhibin beta E |
| chr10_+_76531593 | 0.53 |
ENSMUST00000048678.6
|
Lss
|
lanosterol synthase |
| chr4_-_106492283 | 0.52 |
ENSMUST00000054472.3
|
Bsnd
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr6_-_85933379 | 0.50 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr17_+_34898931 | 0.50 |
ENSMUST00000097342.3
ENSMUST00000013931.5 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr12_+_8771317 | 0.49 |
ENSMUST00000020911.7
|
Sdc1
|
syndecan 1 |
| chr19_+_6497772 | 0.49 |
ENSMUST00000113458.1
ENSMUST00000113459.1 |
Nrxn2
|
neurexin II |
| chr12_-_79296266 | 0.48 |
ENSMUST00000021547.6
|
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr2_+_30416096 | 0.47 |
ENSMUST00000113601.3
ENSMUST00000113603.3 |
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr11_+_85832551 | 0.47 |
ENSMUST00000000095.6
|
Tbx2
|
T-box 2 |
| chr8_+_105264648 | 0.46 |
ENSMUST00000036221.5
|
Fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr4_-_114908892 | 0.45 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr7_-_63938862 | 0.45 |
ENSMUST00000063694.8
|
Klf13
|
Kruppel-like factor 13 |
| chr5_+_150952607 | 0.45 |
ENSMUST00000078856.6
|
Kl
|
klotho |
| chr7_+_68275970 | 0.45 |
ENSMUST00000153805.1
|
Fam169b
|
family with sequence similarity 169, member B |
| chr2_-_36105271 | 0.43 |
ENSMUST00000112960.1
ENSMUST00000112967.5 ENSMUST00000112963.1 |
Lhx6
|
LIM homeobox protein 6 |
| chr2_-_3419066 | 0.43 |
ENSMUST00000115082.3
|
Meig1
|
meiosis expressed gene 1 |
| chr12_-_104865076 | 0.43 |
ENSMUST00000109937.1
ENSMUST00000109936.1 |
Clmn
|
calmin |
| chr13_-_55488038 | 0.42 |
ENSMUST00000109921.2
ENSMUST00000109923.2 ENSMUST00000021950.8 |
Dbn1
|
drebrin 1 |
| chr10_-_80798476 | 0.41 |
ENSMUST00000036805.5
|
Plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr2_+_74691090 | 0.41 |
ENSMUST00000061745.3
|
Hoxd10
|
homeobox D10 |
| chr9_-_58555129 | 0.41 |
ENSMUST00000165365.1
|
Cd276
|
CD276 antigen |
| chr16_+_30599717 | 0.41 |
ENSMUST00000059078.3
|
Fam43a
|
family with sequence similarity 43, member A |
| chr2_+_30416031 | 0.40 |
ENSMUST00000042055.3
|
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr1_+_135818593 | 0.39 |
ENSMUST00000038760.8
|
Lad1
|
ladinin |
| chr11_+_69632927 | 0.39 |
ENSMUST00000018909.3
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr11_+_70647258 | 0.39 |
ENSMUST00000037534.7
|
Rnf167
|
ring finger protein 167 |
| chr15_-_89355655 | 0.38 |
ENSMUST00000023283.5
|
Lmf2
|
lipase maturation factor 2 |
| chr17_+_23660477 | 0.38 |
ENSMUST00000062967.8
|
Ccdc64b
|
coiled-coil domain containing 64B |
| chr9_+_58134535 | 0.38 |
ENSMUST00000128378.1
ENSMUST00000150820.1 ENSMUST00000167479.1 ENSMUST00000134450.1 |
Stra6
|
stimulated by retinoic acid gene 6 |
| chr12_+_8771405 | 0.38 |
ENSMUST00000171158.1
|
Sdc1
|
syndecan 1 |
| chr4_-_118134869 | 0.37 |
ENSMUST00000097912.1
ENSMUST00000030263.2 ENSMUST00000106410.1 |
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr19_-_7105729 | 0.37 |
ENSMUST00000113383.2
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr6_-_18030435 | 0.37 |
ENSMUST00000010941.2
|
Wnt2
|
wingless-related MMTV integration site 2 |
| chr12_-_57546121 | 0.37 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr4_-_57143437 | 0.37 |
ENSMUST00000095076.3
ENSMUST00000030142.3 |
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b |
| chr17_-_45601915 | 0.37 |
ENSMUST00000169137.1
|
Gm7325
|
predicted gene 7325 |
| chr2_-_168742100 | 0.37 |
ENSMUST00000109177.1
|
Atp9a
|
ATPase, class II, type 9A |
| chr7_-_31054815 | 0.37 |
ENSMUST00000071697.4
ENSMUST00000108110.3 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr3_+_63295815 | 0.36 |
ENSMUST00000029400.1
|
Mme
|
membrane metallo endopeptidase |
| chr8_-_105264586 | 0.36 |
ENSMUST00000034359.3
|
Tradd
|
TNFRSF1A-associated via death domain |
| chr7_-_127930066 | 0.36 |
ENSMUST00000032988.8
|
Prss8
|
protease, serine, 8 (prostasin) |
| chr6_+_5725639 | 0.36 |
ENSMUST00000115556.1
ENSMUST00000115555.1 ENSMUST00000115559.3 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr6_-_38109548 | 0.36 |
ENSMUST00000114908.1
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr3_-_131303144 | 0.36 |
ENSMUST00000106337.2
|
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr11_-_67922136 | 0.35 |
ENSMUST00000021288.3
ENSMUST00000108677.1 |
Usp43
|
ubiquitin specific peptidase 43 |
| chr15_-_50890396 | 0.35 |
ENSMUST00000185183.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr7_-_45896656 | 0.35 |
ENSMUST00000120299.1
|
Syngr4
|
synaptogyrin 4 |
| chr17_-_57247632 | 0.35 |
ENSMUST00000005975.6
|
Gpr108
|
G protein-coupled receptor 108 |
| chr5_-_135962265 | 0.35 |
ENSMUST00000111150.1
|
Srcrb4d
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr17_+_34898463 | 0.34 |
ENSMUST00000114033.2
ENSMUST00000078061.6 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr7_-_45896677 | 0.34 |
ENSMUST00000039049.7
|
Syngr4
|
synaptogyrin 4 |
| chr7_-_100656953 | 0.34 |
ENSMUST00000107046.1
ENSMUST00000107045.1 ENSMUST00000139708.1 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr7_+_144838590 | 0.34 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chr8_+_93810832 | 0.34 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_+_24884651 | 0.34 |
ENSMUST00000153451.2
ENSMUST00000108429.1 |
Rps19
|
ribosomal protein S19 |
| chr11_+_75655873 | 0.33 |
ENSMUST00000108431.2
|
Myo1c
|
myosin IC |
| chr15_+_89355730 | 0.33 |
ENSMUST00000074552.5
ENSMUST00000088717.6 |
Ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr11_-_32222233 | 0.33 |
ENSMUST00000150381.1
ENSMUST00000144902.1 ENSMUST00000020524.8 |
Rhbdf1
|
rhomboid family 1 (Drosophila) |
| chr8_+_123653903 | 0.32 |
ENSMUST00000045487.3
|
Rhou
|
ras homolog gene family, member U |
| chr19_+_25610533 | 0.32 |
ENSMUST00000048935.4
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
| chr8_+_114205590 | 0.32 |
ENSMUST00000049509.6
ENSMUST00000150963.1 |
Vat1l
|
vesicle amine transport protein 1 homolog-like (T. californica) |
| chr4_+_148160613 | 0.32 |
ENSMUST00000047951.8
|
Fbxo2
|
F-box protein 2 |
| chr3_+_146499850 | 0.32 |
ENSMUST00000118280.1
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr5_-_143269958 | 0.31 |
ENSMUST00000161448.1
|
Zfp316
|
zinc finger protein 316 |
| chr15_+_89355716 | 0.31 |
ENSMUST00000036987.5
|
Ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr4_+_108879130 | 0.31 |
ENSMUST00000106651.2
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr17_-_24696147 | 0.31 |
ENSMUST00000046839.8
|
Gfer
|
growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) |
| chr7_+_24884611 | 0.31 |
ENSMUST00000108428.1
|
Rps19
|
ribosomal protein S19 |
| chr6_-_52208694 | 0.31 |
ENSMUST00000062829.7
|
Hoxa6
|
homeobox A6 |
| chr2_+_32535315 | 0.31 |
ENSMUST00000133512.1
ENSMUST00000048375.5 |
Fam102a
|
family with sequence similarity 102, member A |
| chr7_+_25306085 | 0.31 |
ENSMUST00000119703.1
ENSMUST00000108409.1 |
Tmem145
|
transmembrane protein 145 |
| chr17_+_24470393 | 0.30 |
ENSMUST00000053024.6
|
Pgp
|
phosphoglycolate phosphatase |
| chr9_+_114978507 | 0.30 |
ENSMUST00000183104.1
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr10_-_95415283 | 0.30 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr7_-_27355944 | 0.30 |
ENSMUST00000003857.6
|
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr19_+_42090422 | 0.30 |
ENSMUST00000066778.4
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr8_+_14095849 | 0.29 |
ENSMUST00000152652.1
ENSMUST00000133298.1 |
Dlgap2
|
discs, large (Drosophila) homolog-associated protein 2 |
| chr10_+_96616998 | 0.29 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr2_-_32381909 | 0.29 |
ENSMUST00000048792.4
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene |
| chr5_-_137684665 | 0.29 |
ENSMUST00000100544.4
ENSMUST00000031736.9 ENSMUST00000151839.1 |
Agfg2
|
ArfGAP with FG repeats 2 |
| chr13_+_49187485 | 0.29 |
ENSMUST00000049022.8
ENSMUST00000120733.1 |
Ninj1
|
ninjurin 1 |
| chr7_-_29156160 | 0.29 |
ENSMUST00000144795.1
ENSMUST00000134176.1 ENSMUST00000164589.1 ENSMUST00000136256.1 |
Fam98c
|
family with sequence similarity 98, member C |
| chr18_-_74961252 | 0.29 |
ENSMUST00000066532.4
|
Lipg
|
lipase, endothelial |
| chr3_+_90541146 | 0.28 |
ENSMUST00000107333.1
ENSMUST00000107331.1 ENSMUST00000098910.2 |
S100a16
|
S100 calcium binding protein A16 |
| chr4_+_116877376 | 0.27 |
ENSMUST00000044823.3
|
Zswim5
|
zinc finger SWIM-type containing 5 |
| chr9_-_107710475 | 0.27 |
ENSMUST00000080560.3
|
Sema3f
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr4_-_150914401 | 0.27 |
ENSMUST00000105675.1
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr13_-_60177357 | 0.27 |
ENSMUST00000065086.4
|
Gas1
|
growth arrest specific 1 |
| chr11_-_116654245 | 0.27 |
ENSMUST00000021166.5
|
Cygb
|
cytoglobin |
| chr3_-_89393294 | 0.27 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr16_+_93683184 | 0.27 |
ENSMUST00000039620.6
|
Cbr3
|
carbonyl reductase 3 |
| chr1_+_36511867 | 0.27 |
ENSMUST00000001166.7
ENSMUST00000097776.3 |
Cnnm3
|
cyclin M3 |
| chr2_-_54085542 | 0.26 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr15_-_97767644 | 0.26 |
ENSMUST00000128775.2
ENSMUST00000134885.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr10_-_84440591 | 0.26 |
ENSMUST00000020220.8
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr3_+_87948666 | 0.26 |
ENSMUST00000005019.5
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr14_+_62760496 | 0.26 |
ENSMUST00000181344.1
|
4931440J10Rik
|
RIKEN cDNA 4931440J10 gene |
| chr10_-_80399478 | 0.26 |
ENSMUST00000092295.3
ENSMUST00000105349.1 |
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr4_+_139574697 | 0.26 |
ENSMUST00000174078.1
|
Iffo2
|
intermediate filament family orphan 2 |
| chr17_-_70851710 | 0.26 |
ENSMUST00000166395.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr13_+_48261427 | 0.26 |
ENSMUST00000021810.1
|
Id4
|
inhibitor of DNA binding 4 |
| chr4_-_155345696 | 0.26 |
ENSMUST00000103178.4
|
Prkcz
|
protein kinase C, zeta |
| chr18_-_78206408 | 0.26 |
ENSMUST00000163367.1
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr7_-_143502515 | 0.26 |
ENSMUST00000010904.4
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr11_-_118125944 | 0.26 |
ENSMUST00000124164.1
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr6_-_115251839 | 0.26 |
ENSMUST00000032462.6
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr6_+_124931378 | 0.26 |
ENSMUST00000032214.7
ENSMUST00000180095.1 |
Mlf2
|
myeloid leukemia factor 2 |
| chr17_+_70561739 | 0.25 |
ENSMUST00000097288.2
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_-_86683761 | 0.25 |
ENSMUST00000018315.3
|
Vmp1
|
vacuole membrane protein 1 |
| chr9_-_107231816 | 0.25 |
ENSMUST00000044532.4
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr15_+_78428564 | 0.25 |
ENSMUST00000166142.2
ENSMUST00000162517.1 ENSMUST00000089414.4 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr4_-_151861698 | 0.25 |
ENSMUST00000049790.7
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr17_+_47436731 | 0.24 |
ENSMUST00000150819.2
|
AI661453
|
expressed sequence AI661453 |
| chr2_-_30093642 | 0.24 |
ENSMUST00000102865.4
|
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr7_+_16309577 | 0.24 |
ENSMUST00000002152.6
|
Bbc3
|
BCL2 binding component 3 |
| chr1_+_191098414 | 0.24 |
ENSMUST00000027943.4
|
Batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr11_-_69579320 | 0.24 |
ENSMUST00000048139.5
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr3_+_146500071 | 0.24 |
ENSMUST00000119130.1
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr7_-_43489967 | 0.24 |
ENSMUST00000107974.1
|
Iglon5
|
IgLON family member 5 |
| chrX_-_73921917 | 0.24 |
ENSMUST00000114389.3
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr11_+_121702591 | 0.24 |
ENSMUST00000125580.1
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr11_+_96316684 | 0.24 |
ENSMUST00000049241.7
|
Hoxb4
|
homeobox B4 |
| chr11_+_114851814 | 0.23 |
ENSMUST00000053361.5
ENSMUST00000021071.7 ENSMUST00000136785.1 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr9_-_45936049 | 0.23 |
ENSMUST00000034590.2
|
Tagln
|
transgelin |
| chr12_-_25096080 | 0.23 |
ENSMUST00000020974.6
|
Id2
|
inhibitor of DNA binding 2 |
| chr17_-_27820445 | 0.23 |
ENSMUST00000114859.1
|
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr11_-_101466222 | 0.23 |
ENSMUST00000040430.7
|
Vat1
|
vesicle amine transport protein 1 homolog (T californica) |
| chr4_+_150914562 | 0.23 |
ENSMUST00000135169.1
|
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr13_+_99344775 | 0.23 |
ENSMUST00000052249.5
|
Mrps27
|
mitochondrial ribosomal protein S27 |
| chr5_-_31047998 | 0.22 |
ENSMUST00000114665.1
ENSMUST00000006817.4 |
Slc5a6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
| chr9_-_119093468 | 0.22 |
ENSMUST00000010804.2
|
Plcd1
|
phospholipase C, delta 1 |
| chr7_+_110627650 | 0.22 |
ENSMUST00000033054.8
|
Adm
|
adrenomedullin |
| chr17_+_46646225 | 0.22 |
ENSMUST00000002844.7
ENSMUST00000113429.1 ENSMUST00000113430.1 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr2_-_160872552 | 0.22 |
ENSMUST00000103111.2
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr8_-_84147858 | 0.22 |
ENSMUST00000117424.2
ENSMUST00000040383.8 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr2_+_136532266 | 0.22 |
ENSMUST00000121717.1
|
Ankef1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr7_-_30973464 | 0.22 |
ENSMUST00000001279.8
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_-_19023538 | 0.22 |
ENSMUST00000036018.5
|
Foxa3
|
forkhead box A3 |
| chr7_+_73391160 | 0.21 |
ENSMUST00000128471.1
|
Rgma
|
RGM domain family, member A |
| chr7_-_99626936 | 0.21 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr15_+_99591028 | 0.21 |
ENSMUST00000169082.1
|
Aqp5
|
aquaporin 5 |
| chr15_+_78406695 | 0.21 |
ENSMUST00000167140.1
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr7_+_29238434 | 0.21 |
ENSMUST00000108237.1
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr15_+_78406777 | 0.21 |
ENSMUST00000169133.1
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr1_+_92831614 | 0.21 |
ENSMUST00000045970.6
|
Gpc1
|
glypican 1 |
| chr17_-_34214459 | 0.21 |
ENSMUST00000121995.1
|
Gm15821
|
predicted gene 15821 |
| chr7_-_93081027 | 0.20 |
ENSMUST00000098303.1
|
Gm9934
|
predicted gene 9934 |
| chr2_-_33431324 | 0.20 |
ENSMUST00000113158.1
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr3_+_146499828 | 0.20 |
ENSMUST00000090031.5
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr17_-_25868727 | 0.20 |
ENSMUST00000026828.5
|
Fam195a
|
family with sequence similarity 195, member A |
| chr4_-_57956283 | 0.20 |
ENSMUST00000030051.5
|
Txn1
|
thioredoxin 1 |
| chr17_-_27820534 | 0.20 |
ENSMUST00000075076.4
ENSMUST00000114863.2 |
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr11_+_100441882 | 0.20 |
ENSMUST00000001599.3
ENSMUST00000107395.2 |
Klhl10
|
kelch-like 10 |
| chr6_-_72958465 | 0.20 |
ENSMUST00000114050.1
|
Tmsb10
|
thymosin, beta 10 |
| chr11_-_102407455 | 0.20 |
ENSMUST00000107098.1
ENSMUST00000018821.2 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr11_+_119942763 | 0.19 |
ENSMUST00000026436.3
ENSMUST00000106231.1 ENSMUST00000075180.5 ENSMUST00000103021.3 ENSMUST00000106233.1 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr2_-_30093607 | 0.19 |
ENSMUST00000081838.6
|
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr8_-_70659645 | 0.19 |
ENSMUST00000070173.7
|
Pgpep1
|
pyroglutamyl-peptidase I |
| chr7_-_19399859 | 0.18 |
ENSMUST00000047170.3
ENSMUST00000108459.2 |
Klc3
|
kinesin light chain 3 |
| chrX_-_73921828 | 0.18 |
ENSMUST00000096316.3
ENSMUST00000114390.1 ENSMUST00000114391.3 ENSMUST00000114387.1 |
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr12_-_79172609 | 0.18 |
ENSMUST00000055262.6
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr19_+_46305682 | 0.18 |
ENSMUST00000111881.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr11_-_62539257 | 0.18 |
ENSMUST00000018653.1
|
Cenpv
|
centromere protein V |
| chr1_+_192190771 | 0.18 |
ENSMUST00000078470.5
ENSMUST00000110844.1 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr2_-_38644087 | 0.18 |
ENSMUST00000028083.5
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7 |
| chr19_+_46003468 | 0.18 |
ENSMUST00000099393.2
|
Hps6
|
Hermansky-Pudlak syndrome 6 |
| chr8_+_123477859 | 0.18 |
ENSMUST00000001520.7
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (yeast) |
| chr12_+_81859964 | 0.18 |
ENSMUST00000021567.5
|
Pcnx
|
pecanex homolog (Drosophila) |
| chr10_-_127666598 | 0.18 |
ENSMUST00000099157.3
|
Nab2
|
Ngfi-A binding protein 2 |
| chr19_+_6975048 | 0.17 |
ENSMUST00000070850.6
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_168431695 | 0.17 |
ENSMUST00000176790.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr11_-_72795801 | 0.17 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chrX_+_161717055 | 0.17 |
ENSMUST00000112338.1
|
Rai2
|
retinoic acid induced 2 |
| chr13_+_73604002 | 0.17 |
ENSMUST00000022102.7
|
Clptm1l
|
CLPTM1-like |
| chr6_-_52260822 | 0.17 |
ENSMUST00000047993.9
|
Hoxa13
|
homeobox A13 |
| chrX_-_73921930 | 0.17 |
ENSMUST00000033763.8
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr11_-_72796028 | 0.17 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0048627 | myoblast development(GO:0048627) |
| 0.3 | 0.8 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.3 | 0.8 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 0.9 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.2 | 0.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.2 | 1.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 0.5 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.2 | 0.9 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.1 | 0.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 1.6 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 0.4 | GO:0090327 | cell communication by chemical coupling(GO:0010643) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.4 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.6 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.1 | 0.4 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.2 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.1 | 0.3 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.6 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:1903168 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.3 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.3 | GO:0097494 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of vesicle size(GO:0097494) |
| 0.1 | 0.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 1.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.6 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.1 | 0.4 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.6 | GO:0070445 | cerebral cortex regionalization(GO:0021796) oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0045578 | bundle of His development(GO:0003166) negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.3 | GO:0097490 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.3 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.3 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.4 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0071163 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1901608 | dense core granule localization(GO:0032253) positive regulation of histone H3-K9 trimethylation(GO:1900114) regulation of vesicle transport along microtubule(GO:1901608) dense core granule transport(GO:1901950) regulation of dense core granule transport(GO:1904809) positive regulation of dense core granule transport(GO:1904811) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.2 | GO:0015669 | gas transport(GO:0015669) carbon dioxide transport(GO:0015670) |
| 0.0 | 0.4 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.3 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.1 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.4 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.4 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.4 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.1 | 0.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0070820 | cytolytic granule(GO:0044194) tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.4 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.2 | GO:0070410 | JUN kinase binding(GO:0008432) co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.7 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0005372 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |