Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
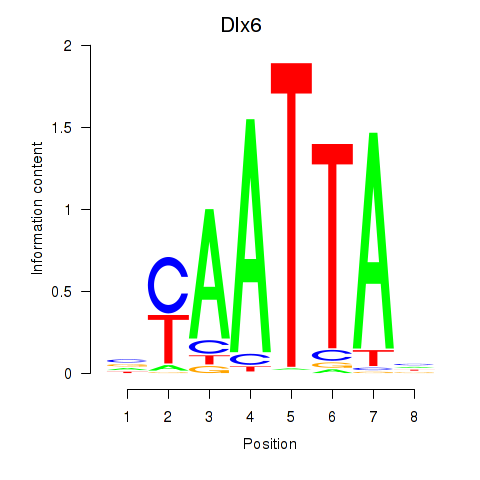
Results for Msx1_Lhx9_Barx1_Rax_Dlx6
Z-value: 0.73
Transcription factors associated with Msx1_Lhx9_Barx1_Rax_Dlx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
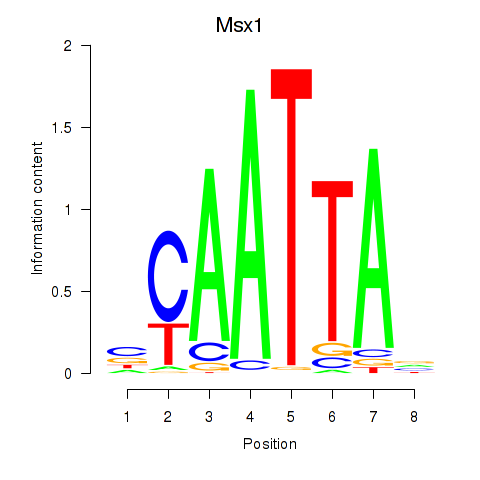
Msx1
|
ENSMUSG00000048450.10 | msh homeobox 1 |
|
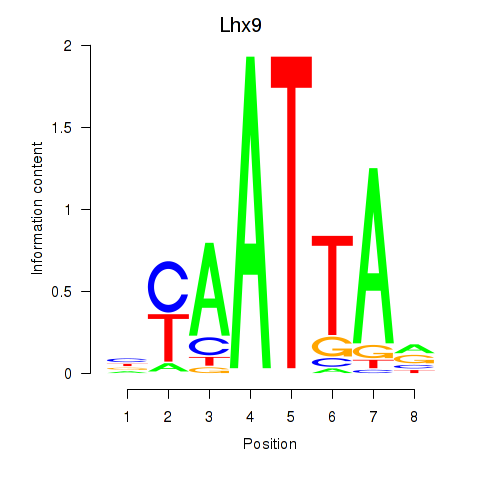
Lhx9
|
ENSMUSG00000019230.8 | LIM homeobox protein 9 |
|
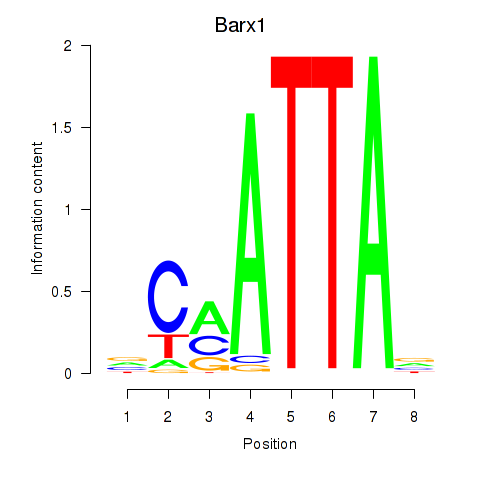
Barx1
|
ENSMUSG00000021381.4 | BarH-like homeobox 1 |
|
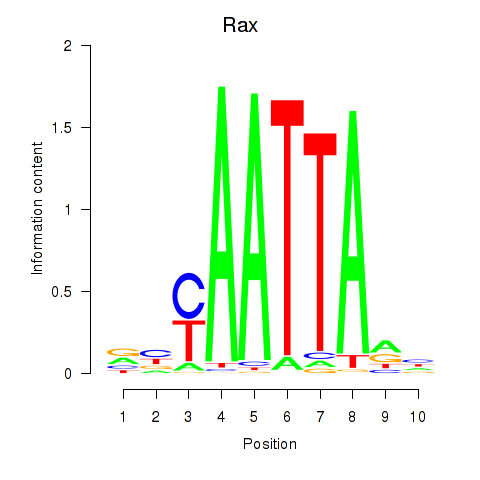
Rax
|
ENSMUSG00000024518.3 | retina and anterior neural fold homeobox |
|
Dlx6
|
ENSMUSG00000029754.7 | distal-less homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barx1 | mm10_v2_chr13_+_48662989_48663046 | 0.47 | 3.9e-03 | Click! |
| Dlx6 | mm10_v2_chr6_+_6863769_6863797 | 0.31 | 6.1e-02 | Click! |
| Rax | mm10_v2_chr18_-_65939048_65939089 | 0.30 | 7.4e-02 | Click! |
| Msx1 | mm10_v2_chr5_-_37824580_37824584 | -0.19 | 2.6e-01 | Click! |
| Lhx9 | mm10_v2_chr1_-_138842429_138842457 | 0.07 | 6.7e-01 | Click! |
Activity profile of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
Sorted Z-values of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_99322943 | 4.50 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr4_+_34893772 | 3.85 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr5_-_134747241 | 2.78 |
ENSMUST00000015138.9
|
Eln
|
elastin |
| chr7_+_126950518 | 2.30 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_-_168767029 | 2.24 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr7_+_126950687 | 2.21 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr14_+_27000362 | 2.01 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_-_79440039 | 2.00 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chr7_+_126950837 | 1.86 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_-_72986716 | 1.56 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr3_-_116253467 | 1.52 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr6_-_116716888 | 1.46 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr9_+_43310763 | 1.45 |
ENSMUST00000034511.5
|
Trim29
|
tripartite motif-containing 29 |
| chr12_-_83487708 | 1.34 |
ENSMUST00000177959.1
ENSMUST00000178756.1 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr12_+_109545390 | 1.32 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr2_+_69219971 | 1.27 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr2_+_9882622 | 1.16 |
ENSMUST00000114919.1
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr16_-_42340595 | 1.12 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr11_+_67200052 | 1.10 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr2_-_168767136 | 1.10 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr10_+_115569986 | 1.08 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr11_+_102604370 | 1.07 |
ENSMUST00000057893.5
|
Fzd2
|
frizzled homolog 2 (Drosophila) |
| chr6_-_136781718 | 1.05 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr2_+_74697663 | 1.05 |
ENSMUST00000059272.8
|
Hoxd9
|
homeobox D9 |
| chr15_-_79285502 | 0.99 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr9_+_43829963 | 0.92 |
ENSMUST00000180221.1
|
Gm3898
|
predicted gene 3898 |
| chr16_-_63864114 | 0.90 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr2_-_116067391 | 0.84 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr13_-_78196373 | 0.84 |
ENSMUST00000125176.2
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr2_+_127909058 | 0.83 |
ENSMUST00000110344.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr15_-_79285470 | 0.79 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr2_+_25372315 | 0.75 |
ENSMUST00000028329.6
ENSMUST00000114293.2 ENSMUST00000100323.2 |
Sapcd2
|
suppressor APC domain containing 2 |
| chr7_-_66427469 | 0.73 |
ENSMUST00000015278.7
|
Aldh1a3
|
aldehyde dehydrogenase family 1, subfamily A3 |
| chr3_-_49757257 | 0.72 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr13_+_55399648 | 0.70 |
ENSMUST00000057167.7
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr18_+_23415400 | 0.70 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr11_-_87359011 | 0.69 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr14_-_110755100 | 0.68 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr6_+_125552948 | 0.67 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr19_-_7966000 | 0.67 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr5_-_53707532 | 0.65 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr17_-_35697971 | 0.65 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr5_+_34999111 | 0.64 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr19_-_55241236 | 0.62 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr5_+_34999046 | 0.62 |
ENSMUST00000114281.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr15_-_13173607 | 0.60 |
ENSMUST00000036439.4
|
Cdh6
|
cadherin 6 |
| chr3_+_68572245 | 0.59 |
ENSMUST00000170788.2
|
Schip1
|
schwannomin interacting protein 1 |
| chr17_-_31198958 | 0.59 |
ENSMUST00000114549.2
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr15_-_34356421 | 0.57 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr17_+_49615104 | 0.56 |
ENSMUST00000162854.1
|
Kif6
|
kinesin family member 6 |
| chrX_-_139871637 | 0.54 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chr11_-_99244058 | 0.53 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr11_+_67798269 | 0.52 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr9_-_40346290 | 0.50 |
ENSMUST00000121357.1
|
Gramd1b
|
GRAM domain containing 1B |
| chrX_-_74246534 | 0.49 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr1_-_4360256 | 0.46 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr5_+_34999070 | 0.45 |
ENSMUST00000114280.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr17_+_27556641 | 0.45 |
ENSMUST00000119486.1
ENSMUST00000118599.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr9_+_45370185 | 0.45 |
ENSMUST00000085939.6
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr7_-_45830776 | 0.45 |
ENSMUST00000107723.2
ENSMUST00000131384.1 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr2_+_38341068 | 0.44 |
ENSMUST00000133661.1
|
Lhx2
|
LIM homeobox protein 2 |
| chr17_+_35533194 | 0.44 |
ENSMUST00000025273.8
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr17_+_27556668 | 0.44 |
ENSMUST00000117254.1
ENSMUST00000118570.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr9_+_119063429 | 0.44 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr2_+_152754156 | 0.43 |
ENSMUST00000010020.5
|
Cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr17_+_27556613 | 0.42 |
ENSMUST00000117600.1
ENSMUST00000114888.3 |
Hmga1
|
high mobility group AT-hook 1 |
| chr9_+_106368594 | 0.42 |
ENSMUST00000172306.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr9_+_38877126 | 0.42 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr11_-_98053415 | 0.41 |
ENSMUST00000017544.2
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr3_+_159839729 | 0.40 |
ENSMUST00000068952.5
|
Wls
|
wntless homolog (Drosophila) |
| chr10_-_80421847 | 0.39 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr10_-_53647080 | 0.38 |
ENSMUST00000169866.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chr13_-_113663670 | 0.37 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr4_+_5724304 | 0.37 |
ENSMUST00000108380.1
|
Fam110b
|
family with sequence similarity 110, member B |
| chr6_-_30958990 | 0.36 |
ENSMUST00000101589.3
|
Klf14
|
Kruppel-like factor 14 |
| chr6_+_123262107 | 0.36 |
ENSMUST00000032240.2
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr18_-_37969742 | 0.34 |
ENSMUST00000166148.1
ENSMUST00000163131.1 ENSMUST00000043437.7 |
Fchsd1
|
FCH and double SH3 domains 1 |
| chr16_+_45093611 | 0.34 |
ENSMUST00000099498.2
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr3_-_55055038 | 0.32 |
ENSMUST00000029368.2
|
Ccna1
|
cyclin A1 |
| chr17_-_49564262 | 0.31 |
ENSMUST00000057610.6
|
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr16_-_74411292 | 0.31 |
ENSMUST00000117200.1
|
Robo2
|
roundabout homolog 2 (Drosophila) |
| chr12_+_52699297 | 0.31 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr8_+_121116163 | 0.30 |
ENSMUST00000054691.6
|
Foxc2
|
forkhead box C2 |
| chr8_+_4238733 | 0.30 |
ENSMUST00000110998.2
ENSMUST00000062686.4 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr2_-_45117349 | 0.30 |
ENSMUST00000176438.2
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_12119274 | 0.30 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chrX_+_56454871 | 0.30 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr8_-_79248537 | 0.28 |
ENSMUST00000034109.4
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr9_+_96258697 | 0.28 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr17_-_29888570 | 0.27 |
ENSMUST00000171691.1
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr11_+_67200137 | 0.27 |
ENSMUST00000129018.1
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chrX_+_133850980 | 0.26 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr2_+_27165233 | 0.26 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr7_+_43781054 | 0.26 |
ENSMUST00000014058.9
|
Klk10
|
kallikrein related-peptidase 10 |
| chr17_+_46161021 | 0.26 |
ENSMUST00000024748.7
ENSMUST00000172170.1 |
Gtpbp2
|
GTP binding protein 2 |
| chr1_-_52190901 | 0.26 |
ENSMUST00000156887.1
ENSMUST00000129107.1 |
Gls
|
glutaminase |
| chr11_-_119547744 | 0.26 |
ENSMUST00000026670.4
|
Nptx1
|
neuronal pentraxin 1 |
| chr7_+_30763750 | 0.25 |
ENSMUST00000165887.1
ENSMUST00000085691.4 ENSMUST00000085688.4 ENSMUST00000054427.6 |
Dmkn
|
dermokine |
| chr8_+_4238815 | 0.25 |
ENSMUST00000003027.7
ENSMUST00000110999.1 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr6_-_52226165 | 0.25 |
ENSMUST00000114425.2
|
Hoxa9
|
homeobox A9 |
| chr13_-_53473074 | 0.24 |
ENSMUST00000021922.8
|
Msx2
|
msh homeobox 2 |
| chr2_+_130012336 | 0.24 |
ENSMUST00000110299.2
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr15_-_66812593 | 0.23 |
ENSMUST00000100572.3
|
Sla
|
src-like adaptor |
| chr6_-_128275577 | 0.23 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr15_+_101473472 | 0.23 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr12_-_111813834 | 0.22 |
ENSMUST00000021715.5
|
Xrcc3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr5_+_27261916 | 0.22 |
ENSMUST00000101471.3
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr2_+_52038005 | 0.22 |
ENSMUST00000065927.5
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr19_+_24673998 | 0.22 |
ENSMUST00000057243.4
|
Tmem252
|
transmembrane protein 252 |
| chr5_-_5266038 | 0.21 |
ENSMUST00000115451.1
ENSMUST00000115452.1 ENSMUST00000131392.1 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_+_3114220 | 0.21 |
ENSMUST00000072955.5
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chrX_+_159303266 | 0.21 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr18_-_24603464 | 0.20 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr12_+_38781093 | 0.20 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr12_-_104473236 | 0.20 |
ENSMUST00000021513.4
|
Gsc
|
goosecoid homeobox |
| chr2_+_106693185 | 0.20 |
ENSMUST00000111063.1
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr15_+_102966794 | 0.19 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr13_+_118714678 | 0.19 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chr13_-_73678005 | 0.19 |
ENSMUST00000022105.7
ENSMUST00000109680.2 ENSMUST00000109679.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr10_+_88146992 | 0.19 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr17_+_75005523 | 0.19 |
ENSMUST00000001927.5
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr4_-_129558355 | 0.18 |
ENSMUST00000167288.1
ENSMUST00000134336.1 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_129558387 | 0.18 |
ENSMUST00000067240.4
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr6_+_37870786 | 0.17 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr18_-_24603791 | 0.17 |
ENSMUST00000070726.3
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr1_-_172027269 | 0.17 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr15_+_79690869 | 0.16 |
ENSMUST00000046463.8
|
Gtpbp1
|
GTP binding protein 1 |
| chr1_-_172027251 | 0.16 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr6_-_102464667 | 0.16 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr14_-_108914237 | 0.16 |
ENSMUST00000100322.2
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr6_+_6248659 | 0.16 |
ENSMUST00000181633.1
ENSMUST00000176283.1 ENSMUST00000175814.1 ENSMUST00000181192.1 |
Gm20619
|
predicted gene 20619 |
| chr10_+_88885992 | 0.16 |
ENSMUST00000020255.6
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr6_-_147264124 | 0.16 |
ENSMUST00000052296.6
|
Pthlh
|
parathyroid hormone-like peptide |
| chrX_+_166344692 | 0.15 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr1_+_172555932 | 0.15 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr5_-_122989086 | 0.15 |
ENSMUST00000046073.9
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr3_+_55782500 | 0.14 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr3_+_61364507 | 0.14 |
ENSMUST00000049064.2
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chr14_-_37098211 | 0.14 |
ENSMUST00000022337.9
|
Cdhr1
|
cadherin-related family member 1 |
| chr5_-_122989260 | 0.13 |
ENSMUST00000118027.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr9_-_103222063 | 0.13 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr1_+_110099295 | 0.13 |
ENSMUST00000134301.1
|
Cdh7
|
cadherin 7, type 2 |
| chr8_+_83666827 | 0.13 |
ENSMUST00000019608.5
|
Ptger1
|
prostaglandin E receptor 1 (subtype EP1) |
| chr2_+_86007778 | 0.13 |
ENSMUST00000062166.1
|
Olfr1032
|
olfactory receptor 1032 |
| chr10_+_102159000 | 0.12 |
ENSMUST00000020039.6
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_62123106 | 0.12 |
ENSMUST00000034052.6
ENSMUST00000034054.7 |
Anxa10
|
annexin A10 |
| chr1_-_152625212 | 0.12 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr17_+_34039437 | 0.12 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr11_+_52098681 | 0.12 |
ENSMUST00000020608.2
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
| chr8_-_87959560 | 0.12 |
ENSMUST00000109655.2
|
Zfp423
|
zinc finger protein 423 |
| chr13_+_111255010 | 0.12 |
ENSMUST00000054716.3
|
Actbl2
|
actin, beta-like 2 |
| chr8_-_120228221 | 0.12 |
ENSMUST00000183235.1
|
A330074K22Rik
|
RIKEN cDNA A330074K22 gene |
| chr5_+_138187485 | 0.12 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr10_+_99263224 | 0.11 |
ENSMUST00000020118.4
|
Dusp6
|
dual specificity phosphatase 6 |
| chr2_+_71389239 | 0.11 |
ENSMUST00000028408.2
|
Hat1
|
histone aminotransferase 1 |
| chr9_-_79977782 | 0.11 |
ENSMUST00000093811.4
|
Filip1
|
filamin A interacting protein 1 |
| chr9_+_102506072 | 0.11 |
ENSMUST00000039390.4
|
Ky
|
kyphoscoliosis peptidase |
| chr10_+_73821857 | 0.11 |
ENSMUST00000177128.1
ENSMUST00000064562.7 ENSMUST00000129404.2 ENSMUST00000105426.3 ENSMUST00000131321.2 ENSMUST00000126920.2 ENSMUST00000147189.2 ENSMUST00000105424.3 ENSMUST00000092420.6 ENSMUST00000105429.3 ENSMUST00000131724.2 ENSMUST00000152655.2 ENSMUST00000151116.2 ENSMUST00000155701.2 ENSMUST00000152819.2 ENSMUST00000125517.2 ENSMUST00000124046.1 ENSMUST00000149977.2 ENSMUST00000146682.1 ENSMUST00000177107.1 |
Pcdh15
|
protocadherin 15 |
| chr4_-_58206596 | 0.11 |
ENSMUST00000042850.8
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr5_-_122988533 | 0.11 |
ENSMUST00000086200.4
ENSMUST00000156474.1 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_-_97700327 | 0.11 |
ENSMUST00000018681.7
|
Pcgf2
|
polycomb group ring finger 2 |
| chr9_+_17030045 | 0.11 |
ENSMUST00000164523.2
|
Gm5611
|
predicted gene 5611 |
| chr17_+_17402672 | 0.11 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr4_-_58499398 | 0.10 |
ENSMUST00000107570.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr13_-_106847267 | 0.10 |
ENSMUST00000057427.4
|
Lrrc70
|
leucine rich repeat containing 70 |
| chr13_+_55445301 | 0.10 |
ENSMUST00000001115.8
ENSMUST00000099482.3 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr2_+_78051155 | 0.10 |
ENSMUST00000145972.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr2_+_78051220 | 0.10 |
ENSMUST00000144728.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr16_-_95586585 | 0.10 |
ENSMUST00000077773.6
|
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chr2_-_72813665 | 0.09 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr7_-_44986313 | 0.09 |
ENSMUST00000045325.6
ENSMUST00000085387.4 ENSMUST00000107840.1 ENSMUST00000107843.3 ENSMUST00000107842.3 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr6_+_15196949 | 0.09 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr5_-_137531952 | 0.09 |
ENSMUST00000140139.1
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr15_+_21111452 | 0.09 |
ENSMUST00000075132.6
|
Cdh12
|
cadherin 12 |
| chr19_-_5560570 | 0.08 |
ENSMUST00000025861.1
|
Ovol1
|
OVO homolog-like 1 (Drosophila) |
| chr18_-_61707583 | 0.08 |
ENSMUST00000025472.1
|
Pcyox1l
|
prenylcysteine oxidase 1 like |
| chr7_+_101896817 | 0.08 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr6_+_87428986 | 0.08 |
ENSMUST00000032125.5
|
Bmp10
|
bone morphogenetic protein 10 |
| chr10_+_102158858 | 0.08 |
ENSMUST00000138522.1
ENSMUST00000163753.1 ENSMUST00000138016.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr7_+_25681158 | 0.08 |
ENSMUST00000108403.3
|
B9d2
|
B9 protein domain 2 |
| chr18_+_44104407 | 0.08 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr1_-_158356258 | 0.08 |
ENSMUST00000004133.8
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr16_-_97170707 | 0.07 |
ENSMUST00000056102.7
|
Dscam
|
Down syndrome cell adhesion molecule |
| chr7_-_102210120 | 0.07 |
ENSMUST00000070165.5
|
Nup98
|
nucleoporin 98 |
| chr1_-_126738167 | 0.07 |
ENSMUST00000160693.1
|
Nckap5
|
NCK-associated protein 5 |
| chr4_+_21848039 | 0.07 |
ENSMUST00000098238.2
ENSMUST00000108229.1 |
Sfrs18
|
serine/arginine-rich splicing factor 18 |
| chr2_+_153938218 | 0.07 |
ENSMUST00000109757.1
|
Bpifb4
|
BPI fold containing family B, member 4 |
| chr18_+_37819543 | 0.07 |
ENSMUST00000055935.5
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr7_-_81493725 | 0.06 |
ENSMUST00000119121.1
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr1_+_132298606 | 0.06 |
ENSMUST00000046071.4
|
Klhdc8a
|
kelch domain containing 8A |
| chr16_-_29544852 | 0.06 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr3_-_59220150 | 0.06 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr1_+_137928100 | 0.06 |
ENSMUST00000054333.2
|
A130050O07Rik
|
RIKEN cDNA A130050O07 gene |
| chr4_+_19280850 | 0.06 |
ENSMUST00000102999.1
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr15_+_92597104 | 0.06 |
ENSMUST00000035399.8
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr12_+_72441933 | 0.06 |
ENSMUST00000161284.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr7_-_116198487 | 0.05 |
ENSMUST00000181981.1
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr13_+_42680565 | 0.05 |
ENSMUST00000128646.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr8_+_54954728 | 0.05 |
ENSMUST00000033915.7
|
Gpm6a
|
glycoprotein m6a |
| chr4_+_19818722 | 0.05 |
ENSMUST00000035890.7
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx1_Lhx9_Barx1_Rax_Dlx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 1.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.7 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 | 0.9 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 | 3.7 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.2 | 0.7 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.7 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 6.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 2.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.7 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 3.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.2 | 4.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 2.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 1.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 1.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 1.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 2.6 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 1.6 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 2.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.0 | 1.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 3.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 3.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 1.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.7 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 3.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 2.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 3.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 1.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |