Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
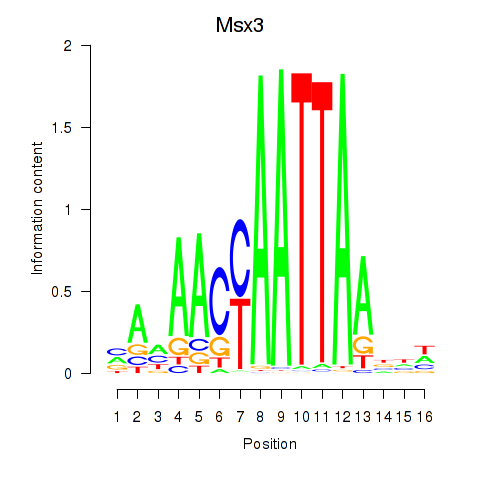
Results for Msx3
Z-value: 0.86
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSMUSG00000025469.9 | msh homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx3 | mm10_v2_chr7_-_140049083_140049089 | 0.32 | 5.6e-02 | Click! |
Activity profile of Msx3 motif
Sorted Z-values of Msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_168767029 | 6.60 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr2_-_168767136 | 5.70 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr14_+_27000362 | 4.15 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_-_52190901 | 2.90 |
ENSMUST00000156887.1
ENSMUST00000129107.1 |
Gls
|
glutaminase |
| chr9_+_96258697 | 2.87 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr10_-_76110956 | 2.65 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr3_-_116253467 | 2.47 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr8_+_57511833 | 2.45 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr12_+_102128718 | 2.00 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr15_-_34356421 | 1.89 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr8_-_24725762 | 1.81 |
ENSMUST00000171438.1
ENSMUST00000171611.1 ENSMUST00000033958.7 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chrX_+_9885622 | 1.77 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr2_-_160619971 | 1.40 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chrX_+_101449078 | 1.38 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr17_-_24527925 | 1.37 |
ENSMUST00000176652.1
|
Traf7
|
TNF receptor-associated factor 7 |
| chr9_-_53975246 | 1.34 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr17_-_24527830 | 1.31 |
ENSMUST00000176353.1
ENSMUST00000176237.1 |
Traf7
|
TNF receptor-associated factor 7 |
| chr16_+_49699198 | 1.26 |
ENSMUST00000046777.4
ENSMUST00000142682.1 |
Ift57
|
intraflagellar transport 57 |
| chr10_+_81257277 | 1.11 |
ENSMUST00000117488.1
ENSMUST00000105328.3 ENSMUST00000121205.1 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr2_-_33087169 | 1.08 |
ENSMUST00000102810.3
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr1_-_172027269 | 1.07 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr10_-_88379051 | 1.06 |
ENSMUST00000138159.1
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr2_-_45117349 | 1.03 |
ENSMUST00000176438.2
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr9_+_72806874 | 0.99 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr7_+_101896817 | 0.89 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr7_+_45621805 | 0.87 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr7_+_101896340 | 0.84 |
ENSMUST00000035395.7
ENSMUST00000106973.1 ENSMUST00000144207.1 |
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr13_-_58354862 | 0.77 |
ENSMUST00000043605.5
|
Kif27
|
kinesin family member 27 |
| chr17_+_56613392 | 0.76 |
ENSMUST00000080492.5
|
Rpl36
|
ribosomal protein L36 |
| chr17_+_21383725 | 0.73 |
ENSMUST00000056107.4
ENSMUST00000162659.1 |
Zfp677
|
zinc finger protein 677 |
| chr5_-_105051047 | 0.72 |
ENSMUST00000112718.4
|
Gbp8
|
guanylate-binding protein 8 |
| chr12_+_71136848 | 0.69 |
ENSMUST00000149564.1
ENSMUST00000045907.8 |
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr2_-_20943413 | 0.67 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr11_+_70018421 | 0.66 |
ENSMUST00000108588.1
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr9_+_35423582 | 0.64 |
ENSMUST00000154652.1
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr19_+_5474681 | 0.62 |
ENSMUST00000165485.1
ENSMUST00000166253.1 ENSMUST00000167371.1 ENSMUST00000167855.1 ENSMUST00000070118.7 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr2_+_163658370 | 0.60 |
ENSMUST00000164399.1
ENSMUST00000064703.6 ENSMUST00000099105.2 ENSMUST00000152418.1 ENSMUST00000126182.1 ENSMUST00000131228.1 |
Pkig
|
protein kinase inhibitor, gamma |
| chr9_+_43310763 | 0.58 |
ENSMUST00000034511.5
|
Trim29
|
tripartite motif-containing 29 |
| chr2_-_33086366 | 0.57 |
ENSMUST00000049618.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr15_+_82252397 | 0.56 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr6_-_135254326 | 0.50 |
ENSMUST00000111911.2
ENSMUST00000111910.2 |
Gsg1
|
germ cell-specific gene 1 |
| chr3_-_41742471 | 0.47 |
ENSMUST00000026866.8
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr11_-_99244058 | 0.44 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr17_+_49615104 | 0.42 |
ENSMUST00000162854.1
|
Kif6
|
kinesin family member 6 |
| chr7_+_27486910 | 0.42 |
ENSMUST00000008528.7
|
Sertad1
|
SERTA domain containing 1 |
| chr9_-_40984460 | 0.42 |
ENSMUST00000180872.1
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr12_+_72441852 | 0.41 |
ENSMUST00000162159.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr1_-_34439672 | 0.40 |
ENSMUST00000042493.8
|
Ccdc115
|
coiled-coil domain containing 115 |
| chr13_-_102958084 | 0.32 |
ENSMUST00000099202.3
ENSMUST00000172264.1 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_+_94739276 | 0.32 |
ENSMUST00000073939.6
ENSMUST00000102798.1 |
Tek
|
endothelial-specific receptor tyrosine kinase |
| chrX_+_133850980 | 0.32 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr4_-_129378116 | 0.31 |
ENSMUST00000030610.2
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr3_+_106034661 | 0.29 |
ENSMUST00000170669.2
|
Gm4540
|
predicted gene 4540 |
| chr11_+_53433299 | 0.29 |
ENSMUST00000018382.6
|
Gdf9
|
growth differentiation factor 9 |
| chr4_-_14621805 | 0.28 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr19_-_11283813 | 0.25 |
ENSMUST00000067673.6
|
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr19_+_29951808 | 0.24 |
ENSMUST00000136850.1
|
Il33
|
interleukin 33 |
| chr9_+_100597686 | 0.22 |
ENSMUST00000124487.1
|
Stag1
|
stromal antigen 1 |
| chr13_+_67863324 | 0.22 |
ENSMUST00000078471.5
|
BC048507
|
cDNA sequence BC048507 |
| chr16_-_29544852 | 0.22 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr12_+_72441933 | 0.21 |
ENSMUST00000161284.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr8_+_107031218 | 0.20 |
ENSMUST00000034388.9
|
Vps4a
|
vacuolar protein sorting 4a (yeast) |
| chr3_+_83055516 | 0.20 |
ENSMUST00000150268.1
ENSMUST00000122128.1 |
Plrg1
|
pleiotropic regulator 1, PRL1 homolog (Arabidopsis) |
| chr2_-_174346712 | 0.18 |
ENSMUST00000168292.1
|
Gm20721
|
predicted gene, 20721 |
| chr3_+_32515295 | 0.13 |
ENSMUST00000029203.7
|
Zfp639
|
zinc finger protein 639 |
| chr4_+_134930898 | 0.12 |
ENSMUST00000030622.2
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr12_-_11208948 | 0.12 |
ENSMUST00000049877.1
|
Msgn1
|
mesogenin 1 |
| chr2_-_150255591 | 0.11 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr1_-_4360256 | 0.09 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr6_-_135254170 | 0.09 |
ENSMUST00000111909.1
|
Gsg1
|
germ cell-specific gene 1 |
| chr1_+_34439851 | 0.06 |
ENSMUST00000027303.7
|
Imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr8_+_66511732 | 0.06 |
ENSMUST00000002025.3
|
Tktl2
|
transketolase-like 2 |
| chr14_+_122034660 | 0.05 |
ENSMUST00000045976.6
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr1_-_127840290 | 0.05 |
ENSMUST00000061512.2
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr3_+_66985680 | 0.04 |
ENSMUST00000065047.6
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_-_116896879 | 0.04 |
ENSMUST00000048229.7
|
Myrfl
|
myelin regulatory factor-like |
| chr3_+_41742615 | 0.01 |
ENSMUST00000146165.1
ENSMUST00000119572.1 ENSMUST00000108065.2 ENSMUST00000120167.1 ENSMUST00000026867.7 ENSMUST00000026868.7 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr14_-_118735143 | 0.01 |
ENSMUST00000184172.1
|
RP24-241I19.1
|
RP24-241I19.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 2.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.5 | 2.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 4.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.4 | 1.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 | 2.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 2.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.6 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 1.7 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.3 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.7 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 1.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 1.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 1.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 12.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 2.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 2.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 2.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.7 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 4.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 2.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 14.1 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |