Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
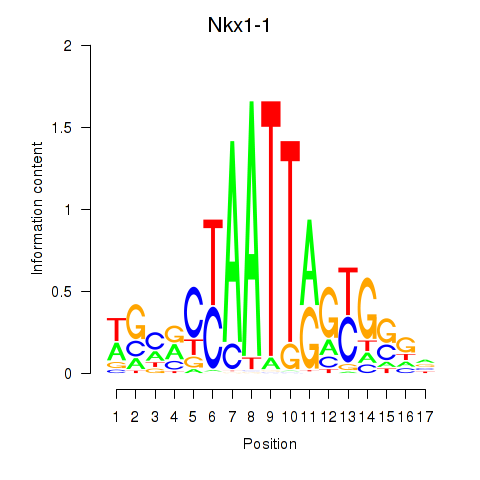
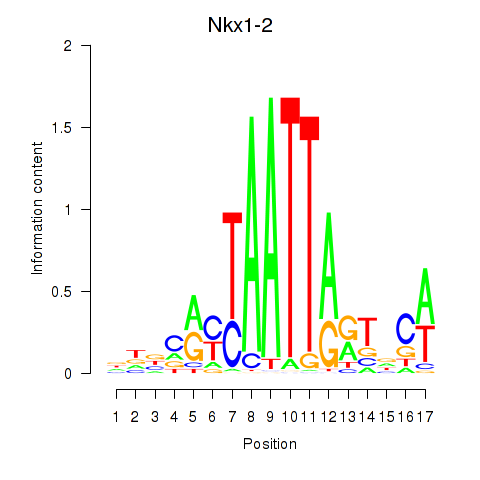
Results for Nkx1-1_Nkx1-2
Z-value: 0.70
Transcription factors associated with Nkx1-1_Nkx1-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx1-1
|
ENSMUSG00000029112.5 | NK1 homeobox 1 |
|
Nkx1-2
|
ENSMUSG00000048528.7 | NK1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx1-1 | mm10_v2_chr5_-_33433976_33433976 | 0.23 | 1.8e-01 | Click! |
| Nkx1-2 | mm10_v2_chr7_-_132599637_132599637 | 0.01 | 9.8e-01 | Click! |
Activity profile of Nkx1-1_Nkx1-2 motif
Sorted Z-values of Nkx1-1_Nkx1-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7966000 | 5.13 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr1_-_150466165 | 2.17 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr10_+_127898515 | 0.93 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr14_-_70429072 | 0.90 |
ENSMUST00000048129.4
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_+_90561102 | 0.85 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr5_+_9100681 | 0.73 |
ENSMUST00000115365.1
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr11_+_43682038 | 0.62 |
ENSMUST00000094294.4
|
Pwwp2a
|
PWWP domain containing 2A |
| chr7_-_73537621 | 0.55 |
ENSMUST00000172704.1
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr11_-_99244058 | 0.53 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr13_+_94083490 | 0.52 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr6_+_37870786 | 0.52 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr4_+_102570065 | 0.52 |
ENSMUST00000097950.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr6_-_57535422 | 0.49 |
ENSMUST00000042766.3
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr10_-_41587753 | 0.48 |
ENSMUST00000160751.1
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr13_-_102905740 | 0.48 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_94372794 | 0.47 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr18_+_4993795 | 0.47 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr11_+_43681998 | 0.46 |
ENSMUST00000061070.5
|
Pwwp2a
|
PWWP domain containing 2A |
| chr8_-_86580664 | 0.45 |
ENSMUST00000131423.1
ENSMUST00000152438.1 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr15_+_9436028 | 0.44 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr1_-_91459254 | 0.43 |
ENSMUST00000069620.8
|
Per2
|
period circadian clock 2 |
| chrX_-_60893430 | 0.40 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr12_-_112802646 | 0.37 |
ENSMUST00000124526.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr19_+_11965817 | 0.37 |
ENSMUST00000025590.9
|
Osbp
|
oxysterol binding protein |
| chr2_-_102400257 | 0.36 |
ENSMUST00000152929.1
|
Trim44
|
tripartite motif-containing 44 |
| chr10_+_116301374 | 0.35 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr16_+_23146536 | 0.34 |
ENSMUST00000023593.5
ENSMUST00000171309.1 |
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr8_+_113635787 | 0.33 |
ENSMUST00000035777.8
|
Mon1b
|
MON1 homolog b (yeast) |
| chr11_-_20332654 | 0.33 |
ENSMUST00000004634.6
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr11_-_21371143 | 0.32 |
ENSMUST00000060895.5
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr11_-_20332689 | 0.32 |
ENSMUST00000109594.1
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr2_-_165388245 | 0.32 |
ENSMUST00000103084.3
|
Zfp334
|
zinc finger protein 334 |
| chr8_+_113635550 | 0.32 |
ENSMUST00000179926.1
|
Mon1b
|
MON1 homolog b (yeast) |
| chr14_-_59365410 | 0.32 |
ENSMUST00000161031.1
ENSMUST00000160425.1 |
Phf11d
|
PHD finger protein 11D |
| chr2_-_69712461 | 0.30 |
ENSMUST00000102706.3
ENSMUST00000073152.6 |
Fastkd1
|
FAST kinase domains 1 |
| chr14_-_59365465 | 0.28 |
ENSMUST00000095157.4
|
Phf11d
|
PHD finger protein 11D |
| chr17_-_34000257 | 0.27 |
ENSMUST00000087189.6
ENSMUST00000173075.1 ENSMUST00000172760.1 ENSMUST00000172912.1 ENSMUST00000025181.10 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr4_-_141599835 | 0.27 |
ENSMUST00000141518.1
ENSMUST00000127455.1 ENSMUST00000105784.1 ENSMUST00000147785.1 |
Fblim1
|
filamin binding LIM protein 1 |
| chr11_-_99851608 | 0.25 |
ENSMUST00000107437.1
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr6_+_68161415 | 0.25 |
ENSMUST00000168090.1
|
Igkv1-115
|
immunoglobulin kappa variable 1-115 |
| chr7_+_51880312 | 0.24 |
ENSMUST00000145049.1
|
Gas2
|
growth arrest specific 2 |
| chr13_-_102906046 | 0.24 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_27195781 | 0.23 |
ENSMUST00000108379.1
ENSMUST00000179391.1 |
BC024978
|
cDNA sequence BC024978 |
| chr2_-_37703845 | 0.22 |
ENSMUST00000155237.1
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chrX_+_20364481 | 0.22 |
ENSMUST00000033372.6
ENSMUST00000115391.1 ENSMUST00000115387.1 |
Rp2h
|
retinitis pigmentosa 2 homolog (human) |
| chr8_-_3694167 | 0.22 |
ENSMUST00000005678.4
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr11_-_100762928 | 0.20 |
ENSMUST00000107360.2
ENSMUST00000055083.3 |
Hcrt
|
hypocretin |
| chr3_+_121953213 | 0.20 |
ENSMUST00000037958.7
ENSMUST00000128366.1 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr8_-_34965631 | 0.19 |
ENSMUST00000033929.4
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr6_-_129775849 | 0.18 |
ENSMUST00000095409.2
|
Gm156
|
predicted gene 156 |
| chr4_+_99030946 | 0.18 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chrX_+_57212110 | 0.18 |
ENSMUST00000033466.1
|
Cd40lg
|
CD40 ligand |
| chr13_-_105054895 | 0.18 |
ENSMUST00000063551.5
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein |
| chr2_-_45112890 | 0.18 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr14_-_8666236 | 0.17 |
ENSMUST00000102996.3
|
4930452B06Rik
|
RIKEN cDNA 4930452B06 gene |
| chr6_-_3399545 | 0.16 |
ENSMUST00000120087.3
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chrX_+_160768013 | 0.15 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr3_+_55782500 | 0.15 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr9_-_54661870 | 0.14 |
ENSMUST00000034822.5
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr18_+_12741324 | 0.14 |
ENSMUST00000115857.2
ENSMUST00000121018.1 ENSMUST00000119108.1 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr16_+_52031549 | 0.14 |
ENSMUST00000114471.1
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr4_+_43493345 | 0.13 |
ENSMUST00000030181.5
ENSMUST00000107922.2 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr7_+_29983948 | 0.13 |
ENSMUST00000148442.1
|
Zfp568
|
zinc finger protein 568 |
| chr5_+_103425181 | 0.13 |
ENSMUST00000048957.9
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr7_-_143460989 | 0.12 |
ENSMUST00000167912.1
ENSMUST00000037287.6 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chrX_-_74085656 | 0.12 |
ENSMUST00000033770.6
|
Mecp2
|
methyl CpG binding protein 2 |
| chr17_+_34931253 | 0.11 |
ENSMUST00000007253.5
|
Neu1
|
neuraminidase 1 |
| chr16_+_44765732 | 0.11 |
ENSMUST00000057488.8
|
Cd200r1
|
CD200 receptor 1 |
| chr6_+_78380700 | 0.11 |
ENSMUST00000101272.1
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr17_-_57031468 | 0.11 |
ENSMUST00000007814.8
|
Khsrp
|
KH-type splicing regulatory protein |
| chr6_+_145615757 | 0.10 |
ENSMUST00000087445.6
|
Tuba3b
|
tubulin, alpha 3B |
| chrX_+_160768179 | 0.10 |
ENSMUST00000112368.2
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chrX_+_107255878 | 0.10 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr10_+_94575257 | 0.10 |
ENSMUST00000121471.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr14_-_50681645 | 0.09 |
ENSMUST00000078075.4
|
Olfr747
|
olfactory receptor 747 |
| chr7_-_19629355 | 0.09 |
ENSMUST00000049912.8
ENSMUST00000094762.3 ENSMUST00000098754.4 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr16_-_16359016 | 0.09 |
ENSMUST00000023477.7
ENSMUST00000096229.3 ENSMUST00000115749.1 |
Dnm1l
|
dynamin 1-like |
| chr10_+_81137953 | 0.09 |
ENSMUST00000117956.1
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr9_-_107872403 | 0.08 |
ENSMUST00000183035.1
|
Rbm6
|
RNA binding motif protein 6 |
| chr15_+_41830921 | 0.08 |
ENSMUST00000166917.1
|
Oxr1
|
oxidation resistance 1 |
| chr5_-_34660068 | 0.08 |
ENSMUST00000041364.9
|
Nop14
|
NOP14 nucleolar protein |
| chr9_-_97369958 | 0.08 |
ENSMUST00000035026.4
|
Trim42
|
tripartite motif-containing 42 |
| chr11_+_99873389 | 0.08 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr8_+_40354303 | 0.08 |
ENSMUST00000136835.1
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr15_+_22549022 | 0.07 |
ENSMUST00000163361.1
|
Cdh18
|
cadherin 18 |
| chr7_-_30552255 | 0.07 |
ENSMUST00000108165.1
ENSMUST00000153594.1 |
BC053749
|
cDNA sequence BC053749 |
| chr9_-_54661666 | 0.07 |
ENSMUST00000128624.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chrX_-_74085586 | 0.07 |
ENSMUST00000123362.1
ENSMUST00000140399.1 ENSMUST00000100750.3 |
Mecp2
|
methyl CpG binding protein 2 |
| chr6_+_145934113 | 0.06 |
ENSMUST00000032383.7
|
Sspn
|
sarcospan |
| chr8_+_83972951 | 0.06 |
ENSMUST00000005606.6
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr11_+_21572221 | 0.05 |
ENSMUST00000020568.3
ENSMUST00000131135.1 |
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr14_+_8666369 | 0.04 |
ENSMUST00000181914.1
ENSMUST00000180444.1 |
4930455B14Rik
|
RIKEN cDNA 4930455B14 gene |
| chr2_+_91265252 | 0.04 |
ENSMUST00000028691.6
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr7_+_24271568 | 0.04 |
ENSMUST00000032696.6
|
Zfp93
|
zinc finger protein 93 |
| chr10_+_116966274 | 0.03 |
ENSMUST00000033651.3
|
D630029K05Rik
|
RIKEN cDNA D630029K05 gene |
| chr1_+_174501796 | 0.03 |
ENSMUST00000030039.7
|
Fmn2
|
formin 2 |
| chr4_-_132463873 | 0.03 |
ENSMUST00000102567.3
|
Med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr3_-_116423930 | 0.03 |
ENSMUST00000106491.2
|
Cdc14a
|
CDC14 cell division cycle 14A |
| chr6_-_52260780 | 0.03 |
ENSMUST00000114416.1
ENSMUST00000147595.3 |
Hoxa13
|
homeobox A13 |
| chr8_-_104534630 | 0.03 |
ENSMUST00000162466.1
ENSMUST00000034349.9 |
Nae1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr17_-_63863791 | 0.03 |
ENSMUST00000050753.3
|
A930002H24Rik
|
RIKEN cDNA A930002H24 gene |
| chr10_+_88147061 | 0.03 |
ENSMUST00000169309.1
|
Nup37
|
nucleoporin 37 |
| chr7_-_24724237 | 0.03 |
ENSMUST00000081657.4
|
Gm4763
|
predicted gene 4763 |
| chr19_+_29367447 | 0.02 |
ENSMUST00000016640.7
|
Cd274
|
CD274 antigen |
| chr3_-_116424007 | 0.02 |
ENSMUST00000090464.4
|
Cdc14a
|
CDC14 cell division cycle 14A |
| chr7_+_3645267 | 0.02 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr7_+_128744870 | 0.02 |
ENSMUST00000042942.8
|
Sec23ip
|
Sec23 interacting protein |
| chr7_-_100964371 | 0.02 |
ENSMUST00000060174.4
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr5_+_104202609 | 0.02 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr9_-_55919605 | 0.01 |
ENSMUST00000037408.8
|
Scaper
|
S phase cyclin A-associated protein in the ER |
| chr17_+_7170101 | 0.01 |
ENSMUST00000024575.6
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr5_+_138187485 | 0.01 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr2_-_132247747 | 0.01 |
ENSMUST00000110163.1
ENSMUST00000180286.1 ENSMUST00000028816.2 |
Tmem230
|
transmembrane protein 230 |
| chr7_+_104003259 | 0.00 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr10_+_88146992 | 0.00 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr5_-_74531619 | 0.00 |
ENSMUST00000113542.2
ENSMUST00000072857.6 ENSMUST00000121330.1 ENSMUST00000151474.1 |
Scfd2
|
Sec1 family domain containing 2 |
| chr12_-_4769217 | 0.00 |
ENSMUST00000053458.5
|
Fam228b
|
family with sequence similarity 228, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx1-1_Nkx1-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 0.9 | GO:0000239 | pachytene(GO:0000239) |
| 0.3 | 2.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 0.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 0.7 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.4 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.3 | GO:2000583 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) metanephric glomerular epithelium development(GO:0072244) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.3 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.2 | GO:1901608 | dense core granule localization(GO:0032253) positive regulation of histone H3-K9 trimethylation(GO:1900114) regulation of vesicle transport along microtubule(GO:1901608) dense core granule transport(GO:1901950) regulation of dense core granule transport(GO:1904809) positive regulation of dense core granule transport(GO:1904811) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.3 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.5 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.4 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.1 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.0 | GO:0070649 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 2.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.7 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0070569 | pyrimidine ribonucleotide binding(GO:0032557) uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |