Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
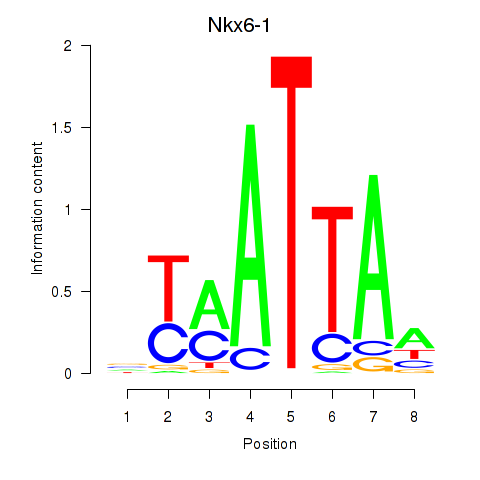
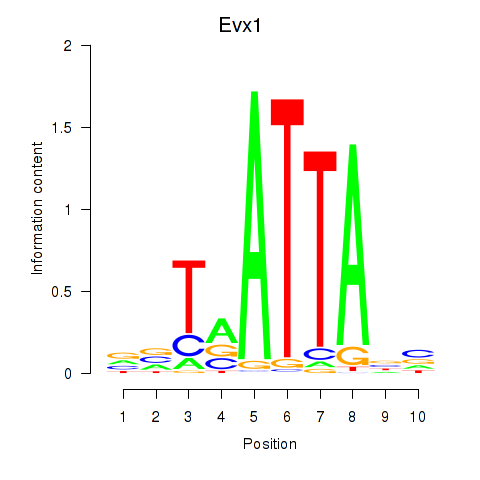
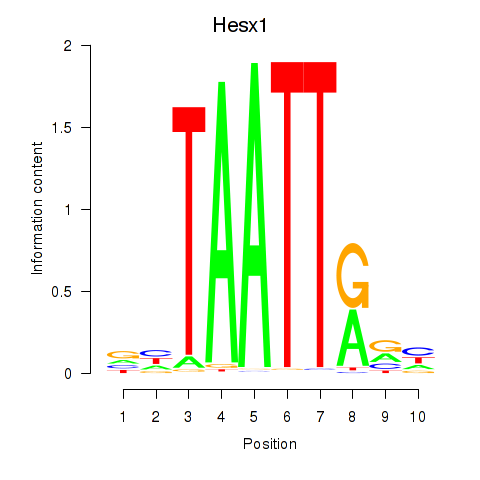
Results for Nkx6-1_Evx1_Hesx1
Z-value: 0.74
Transcription factors associated with Nkx6-1_Evx1_Hesx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-1
|
ENSMUSG00000035187.8 | NK6 homeobox 1 |
|
Evx1
|
ENSMUSG00000005503.8 | even-skipped homeobox 1 |
|
Hesx1
|
ENSMUSG00000040726.8 | homeobox gene expressed in ES cells |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-1 | mm10_v2_chr5_-_101665195_101665226 | 0.30 | 7.2e-02 | Click! |
| Hesx1 | mm10_v2_chr14_+_27000362_27000507 | 0.27 | 1.1e-01 | Click! |
Activity profile of Nkx6-1_Evx1_Hesx1 motif
Sorted Z-values of Nkx6-1_Evx1_Hesx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_62765618 | 4.35 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr10_+_75564086 | 3.05 |
ENSMUST00000141062.1
ENSMUST00000152657.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr3_-_75270073 | 2.95 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr18_+_4993795 | 2.60 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr15_-_101694299 | 2.05 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr14_+_27000362 | 2.01 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr12_+_38780284 | 1.89 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chr3_-_116253467 | 1.79 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr2_+_69897255 | 1.76 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_84734050 | 1.73 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr15_+_25773985 | 1.68 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr10_-_28986280 | 1.60 |
ENSMUST00000152363.1
ENSMUST00000015663.6 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr9_-_123678782 | 1.60 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr17_-_28560704 | 1.60 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chrX_+_56454871 | 1.54 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr5_-_53707532 | 1.45 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr16_-_42340595 | 1.43 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr9_+_43310763 | 1.37 |
ENSMUST00000034511.5
|
Trim29
|
tripartite motif-containing 29 |
| chr15_-_79285502 | 1.35 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr12_+_38780817 | 1.28 |
ENSMUST00000160856.1
|
Etv1
|
ets variant gene 1 |
| chr4_+_8690399 | 1.24 |
ENSMUST00000127476.1
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr8_-_34965631 | 1.24 |
ENSMUST00000033929.4
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr9_-_123678873 | 1.24 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr2_-_168767029 | 1.14 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr13_+_44729535 | 1.10 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr9_+_113812547 | 1.10 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chrM_+_11734 | 1.07 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr18_+_23415400 | 1.06 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr6_+_29853746 | 1.02 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_+_127421208 | 1.01 |
ENSMUST00000168780.1
|
R3hdm2
|
R3H domain containing 2 |
| chr9_-_96719404 | 1.00 |
ENSMUST00000140121.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr8_-_107065632 | 0.99 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr10_+_58394361 | 0.97 |
ENSMUST00000020077.4
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_+_108719649 | 0.94 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr10_+_58394381 | 0.94 |
ENSMUST00000105468.1
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_69219971 | 0.94 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr15_-_100424208 | 0.94 |
ENSMUST00000154331.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr7_+_144838590 | 0.94 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chr1_-_172027251 | 0.91 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr1_-_155417394 | 0.91 |
ENSMUST00000111775.1
ENSMUST00000111774.1 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr13_+_94083490 | 0.89 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chrX_+_159303266 | 0.88 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr15_-_100424092 | 0.87 |
ENSMUST00000154676.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr6_+_121636173 | 0.86 |
ENSMUST00000032203.7
|
A2m
|
alpha-2-macroglobulin |
| chr4_+_103143052 | 0.85 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr19_+_23723279 | 0.85 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr10_+_94575257 | 0.85 |
ENSMUST00000121471.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr6_+_78370877 | 0.81 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chrX_-_164250368 | 0.81 |
ENSMUST00000112263.1
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr13_-_102906046 | 0.79 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_78684262 | 0.78 |
ENSMUST00000145480.1
|
Strn
|
striatin, calmodulin binding protein |
| chr6_-_56923927 | 0.78 |
ENSMUST00000031793.5
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr19_-_55241236 | 0.77 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr6_+_41302265 | 0.77 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr16_+_33794008 | 0.77 |
ENSMUST00000115044.1
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr3_+_122044428 | 0.74 |
ENSMUST00000013995.8
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr18_+_44104407 | 0.72 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr3_-_145032765 | 0.71 |
ENSMUST00000029919.5
|
Clca3
|
chloride channel calcium activated 3 |
| chr17_-_48432723 | 0.71 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr13_+_23544052 | 0.71 |
ENSMUST00000075558.2
|
Hist1h3f
|
histone cluster 1, H3f |
| chr4_-_141846277 | 0.71 |
ENSMUST00000105781.1
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr6_-_41035501 | 0.70 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr8_-_62123106 | 0.69 |
ENSMUST00000034052.6
ENSMUST00000034054.7 |
Anxa10
|
annexin A10 |
| chr1_-_134955847 | 0.69 |
ENSMUST00000168381.1
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr11_-_87359011 | 0.69 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr4_-_141846359 | 0.68 |
ENSMUST00000037059.10
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr8_+_23411490 | 0.68 |
ENSMUST00000033952.7
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr9_+_96258697 | 0.68 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr1_-_174250976 | 0.68 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr3_-_14778452 | 0.67 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr1_-_132390301 | 0.67 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr6_-_41446062 | 0.67 |
ENSMUST00000095999.5
|
Gm10334
|
predicted gene 10334 |
| chr15_-_100425050 | 0.65 |
ENSMUST00000123461.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr1_+_40515362 | 0.65 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr9_-_96719549 | 0.65 |
ENSMUST00000128269.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr15_-_79285470 | 0.64 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr1_+_139454747 | 0.64 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr17_+_3532554 | 0.64 |
ENSMUST00000168560.1
|
Cldn20
|
claudin 20 |
| chr19_-_32196393 | 0.63 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr7_-_28547109 | 0.63 |
ENSMUST00000057974.3
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr4_-_42853888 | 0.62 |
ENSMUST00000107979.1
|
Gm12429
|
predicted gene 12429 |
| chrX_+_49463926 | 0.62 |
ENSMUST00000130558.1
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chrX_+_164090187 | 0.61 |
ENSMUST00000015545.3
|
Tmem27
|
transmembrane protein 27 |
| chr2_-_79456750 | 0.61 |
ENSMUST00000041099.4
|
Neurod1
|
neurogenic differentiation 1 |
| chr5_-_108795352 | 0.61 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr4_+_13743424 | 0.61 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_+_71019548 | 0.60 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr6_+_56924022 | 0.59 |
ENSMUST00000176838.1
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr12_-_55014329 | 0.59 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr1_-_24612700 | 0.58 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr1_-_79440039 | 0.58 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chrX_+_57212110 | 0.58 |
ENSMUST00000033466.1
|
Cd40lg
|
CD40 ligand |
| chrX_-_74246534 | 0.58 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr3_-_130730375 | 0.58 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr3_+_103832562 | 0.57 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr9_-_15357692 | 0.56 |
ENSMUST00000098979.3
ENSMUST00000161132.1 |
5830418K08Rik
|
RIKEN cDNA 5830418K08 gene |
| chr5_-_73191848 | 0.55 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr17_+_71019503 | 0.54 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr14_-_100149764 | 0.52 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr18_-_68429235 | 0.52 |
ENSMUST00000052347.6
|
Mc2r
|
melanocortin 2 receptor |
| chr3_+_64081642 | 0.52 |
ENSMUST00000029406.4
|
Vmn2r1
|
vomeronasal 2, receptor 1 |
| chr2_-_5676046 | 0.51 |
ENSMUST00000114987.3
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chrM_+_9870 | 0.51 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr5_+_34999111 | 0.51 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr6_-_102464667 | 0.50 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr18_-_24603464 | 0.50 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr13_-_102905740 | 0.50 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_124779686 | 0.49 |
ENSMUST00000147669.1
ENSMUST00000128697.1 ENSMUST00000032218.3 ENSMUST00000112475.2 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr2_+_57997884 | 0.49 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr8_+_4238815 | 0.49 |
ENSMUST00000003027.7
ENSMUST00000110999.1 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr4_+_119637704 | 0.49 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr6_-_122340499 | 0.48 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr11_+_67798269 | 0.48 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr10_-_117148474 | 0.48 |
ENSMUST00000020381.3
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr6_+_125552948 | 0.48 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr19_-_11640828 | 0.48 |
ENSMUST00000112984.2
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr16_-_45724600 | 0.48 |
ENSMUST00000096057.4
|
Tagln3
|
transgelin 3 |
| chr8_-_3625274 | 0.48 |
ENSMUST00000004749.6
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr3_-_129755305 | 0.48 |
ENSMUST00000029653.2
|
Egf
|
epidermal growth factor |
| chr2_+_23069210 | 0.47 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chrX_-_9256899 | 0.47 |
ENSMUST00000115553.2
|
Gm14862
|
predicted gene 14862 |
| chr3_+_103832741 | 0.47 |
ENSMUST00000106822.1
|
Bcl2l15
|
BCLl2-like 15 |
| chr4_-_87806296 | 0.47 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_+_125068118 | 0.47 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr17_+_35841668 | 0.47 |
ENSMUST00000174124.1
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr4_-_149126688 | 0.47 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr1_+_88306731 | 0.47 |
ENSMUST00000040210.7
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chrX_+_159697308 | 0.46 |
ENSMUST00000123433.1
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr9_+_54980880 | 0.46 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr7_+_110772604 | 0.44 |
ENSMUST00000005829.6
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_-_66514815 | 0.44 |
ENSMUST00000161879.1
ENSMUST00000159357.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr19_-_56548013 | 0.44 |
ENSMUST00000182059.1
|
Dclre1a
|
DNA cross-link repair 1A, PSO2 homolog (S. cerevisiae) |
| chr16_+_23146536 | 0.44 |
ENSMUST00000023593.5
ENSMUST00000171309.1 |
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr8_-_105568298 | 0.44 |
ENSMUST00000005849.5
|
Agrp
|
agouti related protein |
| chr8_-_24725762 | 0.44 |
ENSMUST00000171438.1
ENSMUST00000171611.1 ENSMUST00000033958.7 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr19_-_34166037 | 0.44 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr15_+_44457522 | 0.43 |
ENSMUST00000166957.1
ENSMUST00000038336.5 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr2_-_7395879 | 0.43 |
ENSMUST00000182404.1
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr6_+_8520008 | 0.43 |
ENSMUST00000162567.1
ENSMUST00000161217.1 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr4_-_126201117 | 0.43 |
ENSMUST00000136157.1
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr14_+_79515618 | 0.43 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr8_-_3624989 | 0.42 |
ENSMUST00000142431.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr2_-_168767136 | 0.42 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr11_-_118103492 | 0.42 |
ENSMUST00000132685.1
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr2_-_116067391 | 0.41 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr4_+_19818722 | 0.41 |
ENSMUST00000035890.7
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr13_-_113663670 | 0.41 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr8_-_3625541 | 0.41 |
ENSMUST00000136105.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr3_+_59952185 | 0.41 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chr3_-_19264959 | 0.41 |
ENSMUST00000121951.1
|
Pde7a
|
phosphodiesterase 7A |
| chr3_-_100489324 | 0.41 |
ENSMUST00000061455.8
|
Fam46c
|
family with sequence similarity 46, member C |
| chr6_+_4755327 | 0.41 |
ENSMUST00000176551.1
|
Peg10
|
paternally expressed 10 |
| chr15_-_34356421 | 0.40 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr12_+_38781093 | 0.40 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr18_-_88927447 | 0.40 |
ENSMUST00000147313.1
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr2_+_4559742 | 0.40 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr6_+_92092369 | 0.40 |
ENSMUST00000113463.1
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr19_-_57197496 | 0.40 |
ENSMUST00000111544.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr12_+_69963452 | 0.39 |
ENSMUST00000110560.1
|
Gm3086
|
predicted gene 3086 |
| chr2_+_110721340 | 0.39 |
ENSMUST00000111016.2
|
Muc15
|
mucin 15 |
| chr5_+_103754560 | 0.39 |
ENSMUST00000153165.1
ENSMUST00000031256.5 |
Aff1
|
AF4/FMR2 family, member 1 |
| chr9_+_32116040 | 0.39 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_+_71909046 | 0.39 |
ENSMUST00000055296.8
|
Polr1a
|
polymerase (RNA) I polypeptide A |
| chr10_+_127420334 | 0.39 |
ENSMUST00000171434.1
|
R3hdm2
|
R3H domain containing 2 |
| chr12_+_109545390 | 0.38 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr12_-_4841583 | 0.38 |
ENSMUST00000020964.5
|
Fkbp1b
|
FK506 binding protein 1b |
| chr6_-_50456085 | 0.38 |
ENSMUST00000146341.1
ENSMUST00000071728.4 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr11_-_62392605 | 0.38 |
ENSMUST00000151498.2
ENSMUST00000159069.1 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr2_+_110721587 | 0.37 |
ENSMUST00000111017.2
|
Muc15
|
mucin 15 |
| chr12_+_81631369 | 0.37 |
ENSMUST00000036116.5
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr5_+_34999070 | 0.37 |
ENSMUST00000114280.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_-_99654983 | 0.37 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr2_-_58160495 | 0.37 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr11_+_23666479 | 0.36 |
ENSMUST00000143117.1
|
Pus10
|
pseudouridylate synthase 10 |
| chr6_-_136781718 | 0.36 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr17_-_40794063 | 0.36 |
ENSMUST00000131699.1
ENSMUST00000024724.7 ENSMUST00000144243.1 |
Crisp2
|
cysteine-rich secretory protein 2 |
| chr12_+_117516479 | 0.36 |
ENSMUST00000109691.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr13_+_23574381 | 0.36 |
ENSMUST00000090776.4
|
Hist1h2ad
|
histone cluster 1, H2ad |
| chr12_-_54986363 | 0.35 |
ENSMUST00000173433.1
ENSMUST00000173803.1 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr2_+_86007778 | 0.35 |
ENSMUST00000062166.1
|
Olfr1032
|
olfactory receptor 1032 |
| chr3_+_66219909 | 0.35 |
ENSMUST00000029421.5
|
Ptx3
|
pentraxin related gene |
| chr9_+_38877126 | 0.35 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr9_-_112187766 | 0.35 |
ENSMUST00000111872.2
ENSMUST00000164754.2 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr5_+_34999046 | 0.34 |
ENSMUST00000114281.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr11_-_77188968 | 0.34 |
ENSMUST00000108400.1
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chrX_-_139871637 | 0.34 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chr11_+_23306910 | 0.34 |
ENSMUST00000137823.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr12_+_31438209 | 0.34 |
ENSMUST00000001254.5
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr15_-_8710734 | 0.34 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_-_37703845 | 0.34 |
ENSMUST00000155237.1
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr17_+_35916977 | 0.33 |
ENSMUST00000151664.1
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr19_+_34100943 | 0.33 |
ENSMUST00000025685.6
|
Lipm
|
lipase, family member M |
| chr2_-_132111440 | 0.33 |
ENSMUST00000128899.1
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr8_-_3625025 | 0.33 |
ENSMUST00000133459.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr5_+_121777929 | 0.33 |
ENSMUST00000160821.1
|
Atxn2
|
ataxin 2 |
| chr10_-_62814539 | 0.33 |
ENSMUST00000173087.1
ENSMUST00000174121.1 |
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr13_-_62607499 | 0.33 |
ENSMUST00000091563.4
|
6720489N17Rik
|
RIKEN cDNA 6720489N17 gene |
| chr2_-_45112890 | 0.33 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_126776939 | 0.32 |
ENSMUST00000038614.5
ENSMUST00000170882.1 ENSMUST00000106359.1 ENSMUST00000106357.1 ENSMUST00000145762.1 ENSMUST00000132643.1 ENSMUST00000106356.1 |
Ypel3
|
yippee-like 3 (Drosophila) |
| chr9_+_13621646 | 0.32 |
ENSMUST00000034401.8
|
Maml2
|
mastermind like 2 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-1_Evx1_Hesx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.5 | 2.5 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.5 | 1.4 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 1.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 1.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 1.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.4 | 3.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 1.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 | 0.7 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.3 | 1.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 3.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 0.9 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 1.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 1.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 3.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 0.6 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.2 | 0.6 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 0.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.2 | 1.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 0.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.9 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.5 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.3 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 | 1.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 1.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.4 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.8 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 2.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.1 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.2 | GO:0061738 | mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 1.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:1900239 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.1 | 0.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.3 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.2 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.7 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.0 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.2 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.2 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 1.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.4 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.8 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0090260 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.4 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.1 | GO:0035926 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.5 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:1904339 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.0 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 1.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 1.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 2.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 2.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 1.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.9 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 2.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 3.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0070695 | HOPS complex(GO:0030897) FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.3 | 0.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 0.9 | GO:0043120 | interleukin-8 binding(GO:0019959) tumor necrosis factor binding(GO:0043120) |
| 0.3 | 2.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 3.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 4.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.7 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 1.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.5 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 3.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.8 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 3.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.8 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 6.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.1 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |