Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr3c2
Z-value: 0.31
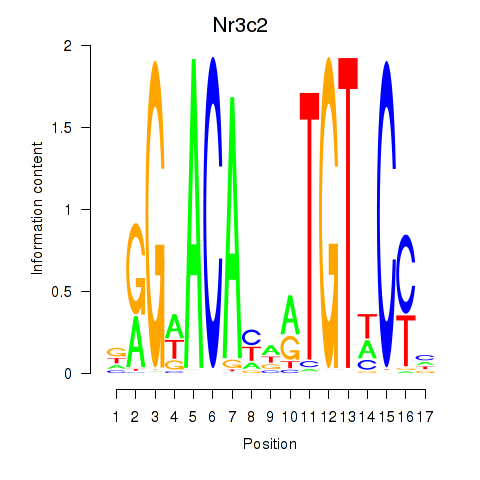
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSMUSG00000031618.7 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | mm10_v2_chr8_+_76902277_76902476 | -0.23 | 1.8e-01 | Click! |
Activity profile of Nr3c2 motif
Sorted Z-values of Nr3c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_125320633 | 0.67 |
ENSMUST00000176655.1
ENSMUST00000176110.1 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr14_-_68533689 | 0.43 |
ENSMUST00000022640.7
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr4_-_49408042 | 0.39 |
ENSMUST00000081541.2
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr3_-_95904683 | 0.37 |
ENSMUST00000147962.1
ENSMUST00000036181.8 |
Car14
|
carbonic anhydrase 14 |
| chr11_+_101448403 | 0.36 |
ENSMUST00000010502.6
|
Ifi35
|
interferon-induced protein 35 |
| chr9_+_123366921 | 0.30 |
ENSMUST00000038863.7
|
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr7_-_25390098 | 0.29 |
ENSMUST00000054301.7
|
Lipe
|
lipase, hormone sensitive |
| chr11_+_98386450 | 0.27 |
ENSMUST00000041301.7
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr15_-_97020322 | 0.25 |
ENSMUST00000166223.1
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr5_+_112288734 | 0.25 |
ENSMUST00000151947.1
|
Tpst2
|
protein-tyrosine sulfotransferase 2 |
| chr14_+_53763083 | 0.21 |
ENSMUST00000180859.1
ENSMUST00000103589.4 |
Trav14-3
|
T cell receptor alpha variable 14-3 |
| chr5_-_24601961 | 0.20 |
ENSMUST00000030791.7
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr1_-_171240055 | 0.20 |
ENSMUST00000131286.1
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr8_+_70697739 | 0.20 |
ENSMUST00000095267.4
|
Jund
|
Jun proto-oncogene related gene d |
| chr15_+_102102926 | 0.18 |
ENSMUST00000169627.1
ENSMUST00000046144.9 |
Tenc1
|
tensin like C1 domain-containing phosphatase |
| chr11_-_75454656 | 0.17 |
ENSMUST00000173320.1
|
Wdr81
|
WD repeat domain 81 |
| chr13_-_119912752 | 0.16 |
ENSMUST00000179502.1
|
Gm21761
|
predicted gene, 21761 |
| chr3_-_88739692 | 0.16 |
ENSMUST00000090946.2
|
Gm10253
|
predicted gene 10253 |
| chr2_-_91444622 | 0.14 |
ENSMUST00000064652.7
ENSMUST00000102594.4 ENSMUST00000094835.2 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr14_+_70530819 | 0.14 |
ENSMUST00000047331.6
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr13_+_22293193 | 0.13 |
ENSMUST00000091735.1
|
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chr6_-_69400097 | 0.13 |
ENSMUST00000177795.1
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr4_+_104913456 | 0.12 |
ENSMUST00000106803.2
ENSMUST00000106804.1 |
1700024P16Rik
|
RIKEN cDNA 1700024P16 gene |
| chr13_+_120173184 | 0.11 |
ENSMUST00000178828.1
|
Gm21818
|
predicted gene, 21818 |
| chr5_-_31697598 | 0.11 |
ENSMUST00000031018.7
|
Rbks
|
ribokinase |
| chr13_+_120317567 | 0.10 |
ENSMUST00000178349.1
|
Tcstv3
|
2-cell-stage, variable group, member 3 |
| chr1_-_157256682 | 0.09 |
ENSMUST00000134543.1
|
Rasal2
|
RAS protein activator like 2 |
| chr6_+_34863130 | 0.08 |
ENSMUST00000074949.3
|
Tmem140
|
transmembrane protein 140 |
| chr5_+_146481943 | 0.06 |
ENSMUST00000179032.1
|
Gm6408
|
predicted gene 6408 |
| chr5_+_31697665 | 0.06 |
ENSMUST00000080598.7
|
Bre
|
brain and reproductive organ-expressed protein |
| chr10_-_14718191 | 0.06 |
ENSMUST00000020016.4
|
Gje1
|
gap junction protein, epsilon 1 |
| chr17_-_23312313 | 0.05 |
ENSMUST00000168033.1
|
Vmn2r114
|
vomeronasal 2, receptor 114 |
| chr13_+_120154627 | 0.05 |
ENSMUST00000179071.1
|
Gm20767
|
predicted gene, 20767 |
| chr9_+_99612468 | 0.04 |
ENSMUST00000042553.7
|
A4gnt
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr13_+_99100698 | 0.04 |
ENSMUST00000181742.1
|
Gm807
|
predicted gene 807 |
| chr9_-_75597643 | 0.04 |
ENSMUST00000164100.1
|
Tmod2
|
tropomodulin 2 |
| chr5_+_123142187 | 0.04 |
ENSMUST00000174836.1
ENSMUST00000163030.2 |
Setd1b
|
SET domain containing 1B |
| chr16_-_96192222 | 0.04 |
ENSMUST00000113804.1
ENSMUST00000054855.7 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chrX_-_102854616 | 0.03 |
ENSMUST00000120314.1
|
Dmrtc1c2
|
DMRT-like family C1c2 |
| chr10_+_21993890 | 0.03 |
ENSMUST00000092673.4
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_82047579 | 0.03 |
ENSMUST00000166578.1
ENSMUST00000080622.7 |
Nhp2l1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr14_-_8309770 | 0.03 |
ENSMUST00000121887.1
ENSMUST00000036070.8 ENSMUST00000137133.1 |
Fam107a
|
family with sequence similarity 107, member A |
| chr1_-_74935549 | 0.02 |
ENSMUST00000094844.3
|
Ccdc108
|
coiled-coil domain containing 108 |
| chrX_+_102802963 | 0.02 |
ENSMUST00000118218.1
|
Dmrtc1c1
|
DMRT-like family C1c1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr3c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.3 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.7 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |