Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
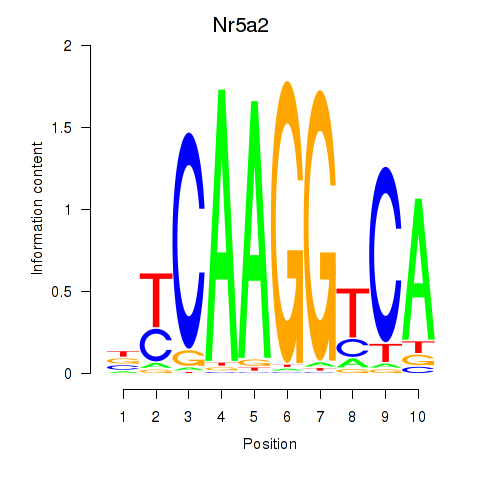
Results for Nr5a2
Z-value: 1.90
Transcription factors associated with Nr5a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr5a2
|
ENSMUSG00000026398.8 | nuclear receptor subfamily 5, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr5a2 | mm10_v2_chr1_-_136953600_136953630 | -0.44 | 7.9e-03 | Click! |
Activity profile of Nr5a2 motif
Sorted Z-values of Nr5a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60582152 | 14.44 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr4_-_61674094 | 13.40 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_61303998 | 12.87 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60421933 | 12.69 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_60662358 | 12.66 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr4_-_60222580 | 12.65 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_61303802 | 12.32 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr4_-_60139857 | 11.86 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr4_-_62054112 | 11.62 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr13_+_21722057 | 5.54 |
ENSMUST00000110476.3
|
Hist1h2bm
|
histone cluster 1, H2bm |
| chr2_+_155611175 | 4.04 |
ENSMUST00000092995.5
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr13_-_56296551 | 3.79 |
ENSMUST00000021970.9
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr2_-_131160006 | 3.48 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr8_+_84970068 | 3.35 |
ENSMUST00000164807.1
|
Prdx2
|
peroxiredoxin 2 |
| chr12_-_110978618 | 3.06 |
ENSMUST00000140788.1
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr12_-_110978981 | 2.97 |
ENSMUST00000135131.1
ENSMUST00000043459.6 ENSMUST00000128353.1 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr14_-_21848924 | 2.90 |
ENSMUST00000124549.1
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr12_-_110978943 | 2.83 |
ENSMUST00000142012.1
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr4_+_63362443 | 2.68 |
ENSMUST00000075341.3
|
Orm2
|
orosomucoid 2 |
| chr12_+_80692591 | 2.50 |
ENSMUST00000140770.1
|
Plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr13_+_21787461 | 2.38 |
ENSMUST00000110473.2
ENSMUST00000102982.1 |
Hist1h2bp
|
histone cluster 1, H2bp |
| chr9_-_106891401 | 2.32 |
ENSMUST00000069036.7
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr13_-_21716143 | 2.32 |
ENSMUST00000091756.1
|
Hist1h2bl
|
histone cluster 1, H2bl |
| chr13_-_21787218 | 2.31 |
ENSMUST00000091751.2
|
Hist1h2an
|
histone cluster 1, H2an |
| chr11_+_74619594 | 2.21 |
ENSMUST00000100866.2
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene |
| chrX_-_141874870 | 2.16 |
ENSMUST00000182079.1
|
Gm15294
|
predicted gene 15294 |
| chr13_-_21753851 | 2.15 |
ENSMUST00000074752.2
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr19_-_46672883 | 2.08 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_+_6415443 | 2.05 |
ENSMUST00000132846.1
|
Ppia
|
peptidylprolyl isomerase A |
| chr15_-_103252810 | 2.04 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr19_+_6400611 | 2.02 |
ENSMUST00000113467.1
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr1_+_92831614 | 2.01 |
ENSMUST00000045970.6
|
Gpc1
|
glypican 1 |
| chr11_-_72266596 | 1.90 |
ENSMUST00000021161.6
ENSMUST00000140167.1 |
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr12_+_105032638 | 1.88 |
ENSMUST00000021522.3
|
Glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr4_+_45184815 | 1.85 |
ENSMUST00000134280.1
ENSMUST00000044773.5 |
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr1_-_167393826 | 1.72 |
ENSMUST00000028005.2
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr1_+_75435930 | 1.71 |
ENSMUST00000037796.7
ENSMUST00000113584.1 ENSMUST00000145166.1 ENSMUST00000143730.1 ENSMUST00000133418.1 ENSMUST00000144874.1 ENSMUST00000140287.1 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr18_+_77185815 | 1.70 |
ENSMUST00000079618.4
|
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr13_+_119623819 | 1.69 |
ENSMUST00000099241.2
|
Ccl28
|
chemokine (C-C motif) ligand 28 |
| chr15_+_85116829 | 1.66 |
ENSMUST00000105085.1
|
Gm10923
|
predicted gene 10923 |
| chr4_+_13751297 | 1.65 |
ENSMUST00000105566.2
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_+_83349446 | 1.61 |
ENSMUST00000136501.1
|
Bola3
|
bolA-like 3 (E. coli) |
| chr2_-_26021679 | 1.46 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr12_-_34291092 | 1.45 |
ENSMUST00000166546.2
|
Gm18025
|
predicted gene, 18025 |
| chr2_-_25571301 | 1.45 |
ENSMUST00000095117.3
ENSMUST00000114223.1 |
Mamdc4
|
MAM domain containing 4 |
| chr15_+_76343504 | 1.44 |
ENSMUST00000023210.6
|
Cyc1
|
cytochrome c-1 |
| chr13_+_119690462 | 1.42 |
ENSMUST00000179869.1
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr4_+_138250403 | 1.41 |
ENSMUST00000105824.1
ENSMUST00000124239.1 ENSMUST00000105818.1 |
Sh2d5
Kif17
|
SH2 domain containing 5 kinesin family member 17 |
| chr6_-_76497631 | 1.41 |
ENSMUST00000097218.5
|
Gm9008
|
predicted pseudogene 9008 |
| chr11_-_102407315 | 1.41 |
ENSMUST00000149777.1
ENSMUST00000154001.1 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr3_+_32736990 | 1.40 |
ENSMUST00000127477.1
ENSMUST00000121778.1 ENSMUST00000154257.1 |
Ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 |
| chr3_-_54915867 | 1.39 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr4_+_138250462 | 1.35 |
ENSMUST00000105823.1
|
Sh2d5
|
SH2 domain containing 5 |
| chr11_-_3504766 | 1.34 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr8_+_84969767 | 1.33 |
ENSMUST00000109733.1
|
Prdx2
|
peroxiredoxin 2 |
| chr2_-_26021532 | 1.32 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr19_-_37178011 | 1.32 |
ENSMUST00000133988.1
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr8_+_84969824 | 1.31 |
ENSMUST00000125893.1
|
Prdx2
|
peroxiredoxin 2 |
| chr1_-_52091066 | 1.31 |
ENSMUST00000105087.1
|
Gm3940
|
predicted gene 3940 |
| chr19_+_6400523 | 1.30 |
ENSMUST00000146831.1
ENSMUST00000035716.8 ENSMUST00000138555.1 ENSMUST00000167240.1 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr15_-_64382908 | 1.29 |
ENSMUST00000177374.1
ENSMUST00000023008.9 ENSMUST00000110115.2 ENSMUST00000110114.3 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr1_+_45981548 | 1.29 |
ENSMUST00000085632.2
|
Rpl23a-ps1
|
ribosomal protein 23A, pseudogene 1 |
| chr18_+_6332587 | 1.29 |
ENSMUST00000097682.2
|
Rpl27-ps3
|
ribosomal protein L27, pseudogene 3 |
| chr17_+_24470393 | 1.28 |
ENSMUST00000053024.6
|
Pgp
|
phosphoglycolate phosphatase |
| chr15_+_98108465 | 1.28 |
ENSMUST00000051226.6
|
Pfkm
|
phosphofructokinase, muscle |
| chr3_-_108085346 | 1.27 |
ENSMUST00000078912.5
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr12_-_71136611 | 1.25 |
ENSMUST00000021486.8
ENSMUST00000166120.1 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr3_+_96576984 | 1.23 |
ENSMUST00000148290.1
|
Gm16253
|
predicted gene 16253 |
| chr12_-_16589743 | 1.23 |
ENSMUST00000111067.2
ENSMUST00000067124.5 |
Lpin1
|
lipin 1 |
| chr10_+_94198955 | 1.21 |
ENSMUST00000020209.9
ENSMUST00000179990.1 |
Ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr14_+_75455957 | 1.21 |
ENSMUST00000164848.1
|
Siah3
|
seven in absentia homolog 3 (Drosophila) |
| chrX_+_71556874 | 1.20 |
ENSMUST00000123100.1
|
Hmgb3
|
high mobility group box 3 |
| chr19_+_6399746 | 1.19 |
ENSMUST00000113468.1
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr19_+_6399857 | 1.19 |
ENSMUST00000146601.1
ENSMUST00000150713.1 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chrX_+_75382384 | 1.19 |
ENSMUST00000033541.4
|
Fundc2
|
FUN14 domain containing 2 |
| chr14_-_77252327 | 1.18 |
ENSMUST00000099431.4
|
Gm10132
|
predicted gene 10132 |
| chr13_+_22043189 | 1.17 |
ENSMUST00000110452.1
|
Hist1h2bj
|
histone cluster 1, H2bj |
| chr6_-_125165576 | 1.17 |
ENSMUST00000183272.1
ENSMUST00000182052.1 ENSMUST00000182277.1 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr2_+_102706356 | 1.16 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr9_-_106891870 | 1.16 |
ENSMUST00000160503.1
ENSMUST00000159620.2 ENSMUST00000160978.1 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr19_-_60874526 | 1.16 |
ENSMUST00000025961.6
|
Prdx3
|
peroxiredoxin 3 |
| chr7_+_30184160 | 1.15 |
ENSMUST00000098594.2
|
Cox7a1
|
cytochrome c oxidase subunit VIIa 1 |
| chr4_-_148159571 | 1.13 |
ENSMUST00000167160.1
ENSMUST00000151246.1 |
Fbxo44
|
F-box protein 44 |
| chr4_-_129227883 | 1.09 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr14_+_34170640 | 1.08 |
ENSMUST00000104925.3
|
Rpl23a-ps3
|
ribosomal protein L23A, pseudogene 3 |
| chr6_+_29853746 | 1.08 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr17_-_83631892 | 1.06 |
ENSMUST00000051482.1
|
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr3_-_96220880 | 1.06 |
ENSMUST00000090782.3
|
Hist2h2ac
|
histone cluster 2, H2ac |
| chr6_+_71199827 | 1.06 |
ENSMUST00000067492.7
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr11_-_82871133 | 1.06 |
ENSMUST00000071152.7
ENSMUST00000108173.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr11_+_31872100 | 1.05 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr8_+_71464910 | 1.04 |
ENSMUST00000048914.6
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr4_+_156215920 | 1.04 |
ENSMUST00000105572.1
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene |
| chr5_+_118065360 | 1.03 |
ENSMUST00000031305.3
|
Gm9754
|
predicted gene 9754 |
| chr8_-_94838255 | 1.03 |
ENSMUST00000161762.1
ENSMUST00000162538.1 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr10_+_80805233 | 1.01 |
ENSMUST00000036016.4
|
Amh
|
anti-Mullerian hormone |
| chr1_+_91250482 | 1.01 |
ENSMUST00000171112.1
|
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr9_+_119937606 | 1.01 |
ENSMUST00000035100.5
|
Ttc21a
|
tetratricopeptide repeat domain 21A |
| chr19_+_21272276 | 1.00 |
ENSMUST00000025659.4
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr11_+_54304005 | 1.00 |
ENSMUST00000000145.5
ENSMUST00000138515.1 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_-_110769345 | 1.00 |
ENSMUST00000098108.2
|
B430319F04Rik
|
RIKEN cDNA B430319F04 gene |
| chr7_-_98145472 | 0.98 |
ENSMUST00000098281.2
|
Omp
|
olfactory marker protein |
| chr10_-_43540945 | 0.98 |
ENSMUST00000147196.1
ENSMUST00000019932.3 |
1700021F05Rik
|
RIKEN cDNA 1700021F05 gene |
| chr2_+_103970115 | 0.97 |
ENSMUST00000111143.1
ENSMUST00000138815.1 |
Lmo2
|
LIM domain only 2 |
| chr13_+_23746734 | 0.97 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr8_+_94152607 | 0.97 |
ENSMUST00000034211.8
|
Mt3
|
metallothionein 3 |
| chr16_+_35770382 | 0.96 |
ENSMUST00000023555.4
|
Hspbap1
|
Hspb associated protein 1 |
| chr10_+_118860826 | 0.96 |
ENSMUST00000059966.4
|
4932442E05Rik
|
RIKEN cDNA 4932442E05 gene |
| chr2_+_30416096 | 0.95 |
ENSMUST00000113601.3
ENSMUST00000113603.3 |
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr7_+_17150210 | 0.94 |
ENSMUST00000108491.1
ENSMUST00000065540.5 |
Ceacam3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr2_+_30416031 | 0.94 |
ENSMUST00000042055.3
|
Ppp2r4
|
protein phosphatase 2A, regulatory subunit B (PR 53) |
| chr17_-_34804546 | 0.93 |
ENSMUST00000025223.8
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr2_+_103970221 | 0.93 |
ENSMUST00000111140.2
ENSMUST00000111139.2 |
Lmo2
|
LIM domain only 2 |
| chr13_-_22042949 | 0.92 |
ENSMUST00000091741.4
|
Hist1h2ag
|
histone cluster 1, H2ag |
| chr15_-_44428303 | 0.92 |
ENSMUST00000038719.6
|
Nudcd1
|
NudC domain containing 1 |
| chr8_-_70523085 | 0.92 |
ENSMUST00000137610.1
ENSMUST00000121623.1 ENSMUST00000093456.5 ENSMUST00000118850.1 |
Kxd1
|
KxDL motif containing 1 |
| chr4_-_73950834 | 0.92 |
ENSMUST00000095023.1
ENSMUST00000030101.3 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr3_-_84040151 | 0.92 |
ENSMUST00000052342.7
|
D930015E06Rik
|
RIKEN cDNA D930015E06 gene |
| chr2_+_32587057 | 0.91 |
ENSMUST00000102818.4
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr7_+_4740111 | 0.91 |
ENSMUST00000098853.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr3_-_86920830 | 0.90 |
ENSMUST00000029719.8
|
Dclk2
|
doublecortin-like kinase 2 |
| chr13_-_74350206 | 0.90 |
ENSMUST00000022062.7
|
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr17_-_45474839 | 0.90 |
ENSMUST00000024731.8
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr7_-_141429351 | 0.89 |
ENSMUST00000164387.1
ENSMUST00000137488.1 ENSMUST00000084436.3 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr13_+_55445301 | 0.89 |
ENSMUST00000001115.8
ENSMUST00000099482.3 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr10_+_14523062 | 0.89 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr4_-_45108038 | 0.88 |
ENSMUST00000107809.2
ENSMUST00000107808.2 ENSMUST00000107807.1 ENSMUST00000107810.2 |
Tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr9_+_109832749 | 0.88 |
ENSMUST00000147777.1
ENSMUST00000035053.5 ENSMUST00000133483.1 |
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr2_+_130576170 | 0.88 |
ENSMUST00000028764.5
|
Oxt
|
oxytocin |
| chr9_+_109832998 | 0.87 |
ENSMUST00000119376.1
ENSMUST00000122343.1 |
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr11_-_120630516 | 0.86 |
ENSMUST00000106181.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr4_-_83324135 | 0.86 |
ENSMUST00000030205.7
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chrX_+_160768013 | 0.86 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr8_-_70120816 | 0.86 |
ENSMUST00000002412.8
|
Ncan
|
neurocan |
| chr2_+_178118975 | 0.85 |
ENSMUST00000108917.1
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr2_+_137663424 | 0.85 |
ENSMUST00000134833.1
|
Gm14064
|
predicted gene 14064 |
| chr4_-_137409777 | 0.84 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr15_-_98728120 | 0.83 |
ENSMUST00000003445.6
|
Fkbp11
|
FK506 binding protein 11 |
| chr7_+_78783119 | 0.83 |
ENSMUST00000032840.4
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr19_+_5878622 | 0.83 |
ENSMUST00000136833.1
ENSMUST00000141362.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr15_-_102516806 | 0.83 |
ENSMUST00000169162.1
ENSMUST00000023812.2 ENSMUST00000165174.1 ENSMUST00000169367.1 ENSMUST00000169377.1 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr7_-_105574324 | 0.82 |
ENSMUST00000081165.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr19_-_6996025 | 0.81 |
ENSMUST00000041686.3
ENSMUST00000180765.1 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr15_-_98221056 | 0.80 |
ENSMUST00000170618.1
ENSMUST00000141911.1 |
Olfr287
|
olfactory receptor 287 |
| chr2_-_73911323 | 0.80 |
ENSMUST00000111996.1
ENSMUST00000018914.2 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr14_+_20929416 | 0.80 |
ENSMUST00000022369.7
|
Vcl
|
vinculin |
| chr10_-_62507737 | 0.79 |
ENSMUST00000020271.6
|
Srgn
|
serglycin |
| chr1_+_135799833 | 0.79 |
ENSMUST00000148201.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr11_+_67586520 | 0.78 |
ENSMUST00000108682.2
|
Gas7
|
growth arrest specific 7 |
| chr11_+_90638127 | 0.77 |
ENSMUST00000020851.8
|
Cox11
|
cytochrome c oxidase assembly protein 11 |
| chr7_+_142434977 | 0.77 |
ENSMUST00000118276.1
ENSMUST00000105976.1 ENSMUST00000097939.2 |
Syt8
|
synaptotagmin VIII |
| chr10_+_128083273 | 0.76 |
ENSMUST00000026459.5
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr15_-_52478228 | 0.75 |
ENSMUST00000081993.1
|
Gm10020
|
predicted pseudogene 10020 |
| chr18_+_77938452 | 0.75 |
ENSMUST00000044622.5
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr4_+_107830958 | 0.75 |
ENSMUST00000106731.2
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr2_+_152105722 | 0.75 |
ENSMUST00000099225.2
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr1_+_36691487 | 0.75 |
ENSMUST00000081180.4
|
Cox5b
|
cytochrome c oxidase subunit Vb |
| chr19_-_7217549 | 0.74 |
ENSMUST00000039758.4
|
Cox8a
|
cytochrome c oxidase subunit VIIIa |
| chr7_+_4740178 | 0.74 |
ENSMUST00000108583.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr13_+_113209659 | 0.74 |
ENSMUST00000038144.8
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr2_+_160888101 | 0.74 |
ENSMUST00000109455.2
ENSMUST00000040872.6 |
Lpin3
|
lipin 3 |
| chr12_+_102554966 | 0.74 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr12_+_12911986 | 0.73 |
ENSMUST00000085720.1
|
Rpl36-ps3
|
ribosomal protein L36, pseudogene 3 |
| chr8_-_84969740 | 0.73 |
ENSMUST00000109736.2
ENSMUST00000140561.1 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr7_+_100006404 | 0.72 |
ENSMUST00000032977.4
|
Chrdl2
|
chordin-like 2 |
| chr1_-_136131171 | 0.72 |
ENSMUST00000146091.3
ENSMUST00000165464.1 ENSMUST00000166747.1 ENSMUST00000134998.1 |
Gm15850
|
predicted gene 15850 |
| chr10_-_80855187 | 0.72 |
ENSMUST00000035775.8
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_-_33276334 | 0.71 |
ENSMUST00000183831.1
|
Gm12117
|
predicted gene 12117 |
| chr8_+_105701624 | 0.71 |
ENSMUST00000093195.6
|
Pard6a
|
par-6 (partitioning defective 6,) homolog alpha (C. elegans) |
| chr7_+_4740137 | 0.70 |
ENSMUST00000130215.1
ENSMUST00000108582.3 |
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr2_-_103073335 | 0.70 |
ENSMUST00000132449.1
ENSMUST00000111183.1 ENSMUST00000011058.2 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr8_+_120668308 | 0.69 |
ENSMUST00000181795.1
|
Cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr15_+_74563738 | 0.69 |
ENSMUST00000170845.1
|
Bai1
|
brain-specific angiogenesis inhibitor 1 |
| chr13_+_21716385 | 0.69 |
ENSMUST00000070124.3
|
Hist1h2ai
|
histone cluster 1, H2ai |
| chr15_-_82764176 | 0.68 |
ENSMUST00000055721.4
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr10_-_62527413 | 0.68 |
ENSMUST00000160643.1
|
Srgn
|
serglycin |
| chr2_+_160888156 | 0.68 |
ENSMUST00000109457.2
|
Lpin3
|
lipin 3 |
| chr10_-_62508097 | 0.68 |
ENSMUST00000159020.1
|
Srgn
|
serglycin |
| chr7_+_100494044 | 0.67 |
ENSMUST00000153287.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_+_67586675 | 0.67 |
ENSMUST00000108680.1
|
Gas7
|
growth arrest specific 7 |
| chr11_+_101078411 | 0.66 |
ENSMUST00000019445.5
|
Hsd17b1
|
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr7_+_100493795 | 0.66 |
ENSMUST00000129324.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_-_5065276 | 0.66 |
ENSMUST00000109895.1
ENSMUST00000152257.1 ENSMUST00000037146.3 ENSMUST00000056649.6 |
Gas2l1
|
growth arrest-specific 2 like 1 |
| chr2_+_140170641 | 0.65 |
ENSMUST00000044825.4
|
Ndufaf5
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
| chr9_-_77347816 | 0.65 |
ENSMUST00000184138.1
ENSMUST00000184006.1 ENSMUST00000185144.1 ENSMUST00000034910.9 |
Mlip
|
muscular LMNA-interacting protein |
| chr2_+_25262589 | 0.65 |
ENSMUST00000114336.3
|
Tprn
|
taperin |
| chr1_-_51941261 | 0.65 |
ENSMUST00000097103.3
|
Gm8420
|
predicted gene 8420 |
| chr1_+_63176818 | 0.65 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr9_+_57910974 | 0.63 |
ENSMUST00000163329.1
|
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr13_+_51100810 | 0.62 |
ENSMUST00000095797.5
|
Spin1
|
spindlin 1 |
| chr12_-_111966954 | 0.62 |
ENSMUST00000021719.5
|
2010107E04Rik
|
RIKEN cDNA 2010107E04 gene |
| chr9_-_77347787 | 0.62 |
ENSMUST00000184848.1
ENSMUST00000184415.1 |
Mlip
|
muscular LMNA-interacting protein |
| chr18_+_37955544 | 0.61 |
ENSMUST00000070709.2
ENSMUST00000177058.1 ENSMUST00000169360.2 ENSMUST00000163591.2 ENSMUST00000091932.5 |
Rell2
|
RELT-like 2 |
| chr5_-_66151903 | 0.60 |
ENSMUST00000167950.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr11_+_53720790 | 0.60 |
ENSMUST00000048605.2
|
Il5
|
interleukin 5 |
| chr4_-_83324239 | 0.60 |
ENSMUST00000048274.4
ENSMUST00000102823.3 |
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr19_-_47090610 | 0.60 |
ENSMUST00000096014.3
|
Usmg5
|
upregulated during skeletal muscle growth 5 |
| chr1_-_4785671 | 0.59 |
ENSMUST00000130201.1
ENSMUST00000156816.1 |
Mrpl15
|
mitochondrial ribosomal protein L15 |
| chr16_+_10545339 | 0.59 |
ENSMUST00000066345.7
ENSMUST00000115824.3 ENSMUST00000155633.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr10_-_75517324 | 0.59 |
ENSMUST00000039796.7
|
Gucd1
|
guanylyl cyclase domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr5a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.6 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.2 | 3.5 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 1.1 | 12.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.9 | 3.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.7 | 2.7 | GO:0033367 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.7 | 2.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.5 | 6.0 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.5 | 1.9 | GO:0015744 | succinate transport(GO:0015744) |
| 0.5 | 1.4 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.4 | 1.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.4 | 1.3 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.4 | 1.3 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.4 | 2.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.4 | 1.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 0.9 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 | 1.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 1.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 1.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 1.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 1.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 1.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 1.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 1.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 1.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.7 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.2 | 1.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 0.7 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.2 | 0.9 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 1.3 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.2 | 0.6 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 2.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 2.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 2.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 0.8 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.2 | 1.0 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 | 1.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.5 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.2 | 0.9 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.1 | 1.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.4 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.8 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.4 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.3 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 1.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.9 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.3 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.1 | 0.4 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.7 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 1.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 9.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.2 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.3 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 0.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 2.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.4 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.2 | GO:1902477 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 3.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 5.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.3 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.5 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 1.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 1.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.6 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.8 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.1 | GO:0006113 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.8 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.7 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 1.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 1.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 2.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.2 | 1.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 0.8 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.2 | 1.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 11.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.5 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 3.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 0.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.1 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 2.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 4.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 7.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.7 | 7.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 1.9 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.5 | 2.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.4 | 2.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 1.2 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.3 | 1.3 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 1.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 2.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 1.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.7 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 0.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 1.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.7 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.2 | 1.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.2 | 4.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 0.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 1.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.4 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 5.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 5.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 2.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.6 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 1.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 1.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 1.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 2.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 6.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.9 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 3.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 6.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 10.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 5.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 11.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.0 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |