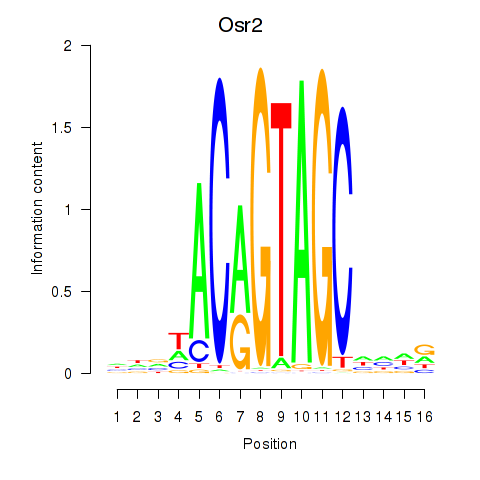
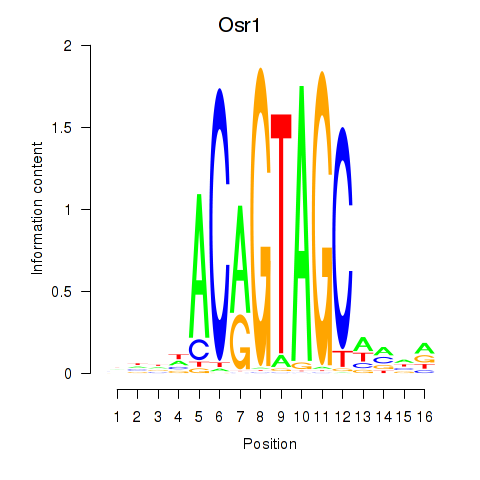
Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Osr2_Osr1
Z-value: 0.42
Transcription factors associated with Osr2_Osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Osr2
|
ENSMUSG00000022330.4 | odd-skipped related 2 |
|
Osr1
|
ENSMUSG00000048387.7 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr2 | mm10_v2_chr15_+_35296090_35296112 | -0.30 | 7.4e-02 | Click! |
| Osr1 | mm10_v2_chr12_+_9574437_9574448 | -0.18 | 2.9e-01 | Click! |
Activity profile of Osr2_Osr1 motif
Sorted Z-values of Osr2_Osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20601958 | 1.03 |
ENSMUST00000087638.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr10_+_84838143 | 0.70 |
ENSMUST00000095388.4
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr8_+_21391811 | 0.65 |
ENSMUST00000120874.3
|
Gm21002
|
predicted gene, 21002 |
| chr8_+_21509258 | 0.60 |
ENSMUST00000084042.3
|
Defa20
|
defensin, alpha, 20 |
| chr8_+_21162277 | 0.59 |
ENSMUST00000098897.1
|
Defa22
|
defensin, alpha, 22 |
| chr18_+_20944607 | 0.59 |
ENSMUST00000050004.1
|
Rnf125
|
ring finger protein 125 |
| chr16_-_18413452 | 0.54 |
ENSMUST00000165430.1
ENSMUST00000147720.1 |
Comt
|
catechol-O-methyltransferase |
| chr8_+_21378560 | 0.53 |
ENSMUST00000170275.2
|
Defa2
|
defensin, alpha, 2 |
| chr8_+_21529031 | 0.52 |
ENSMUST00000084041.3
|
Gm15308
|
predicted gene 15308 |
| chr3_-_89322883 | 0.49 |
ENSMUST00000029673.5
|
Efna3
|
ephrin A3 |
| chr7_+_119561699 | 0.44 |
ENSMUST00000167935.2
ENSMUST00000130583.1 |
Acsm2
|
acyl-CoA synthetase medium-chain family member 2 |
| chr3_-_98763053 | 0.44 |
ENSMUST00000107019.1
ENSMUST00000107018.1 ENSMUST00000125483.1 ENSMUST00000137008.1 ENSMUST00000135706.1 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr8_+_21025545 | 0.44 |
ENSMUST00000076754.2
|
Defa21
|
defensin, alpha, 21 |
| chr17_-_57228003 | 0.43 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr10_+_77829467 | 0.40 |
ENSMUST00000092368.2
|
Tspear
|
thrombospondin type laminin G domain and EAR repeats |
| chr15_-_60921270 | 0.39 |
ENSMUST00000096418.3
|
A1bg
|
alpha-1-B glycoprotein |
| chr1_+_88138364 | 0.38 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr5_-_145879857 | 0.38 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr11_-_53470479 | 0.37 |
ENSMUST00000057722.2
|
Gm9837
|
predicted gene 9837 |
| chr19_-_6067785 | 0.35 |
ENSMUST00000162575.1
ENSMUST00000159084.1 ENSMUST00000161718.1 ENSMUST00000162810.1 ENSMUST00000025713.5 ENSMUST00000113543.2 ENSMUST00000161528.1 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_-_24109582 | 0.35 |
ENSMUST00000041180.5
|
Taar9
|
trace amine-associated receptor 9 |
| chr1_-_180996145 | 0.35 |
ENSMUST00000154133.1
|
Ephx1
|
epoxide hydrolase 1, microsomal |
| chrY_-_6681243 | 0.34 |
ENSMUST00000115940.1
|
Gm21719
|
predicted gene, 21719 |
| chr17_-_45593626 | 0.34 |
ENSMUST00000163905.1
ENSMUST00000167692.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_-_37769624 | 0.32 |
ENSMUST00000175941.1
|
Zfp536
|
zinc finger protein 536 |
| chr15_+_62178175 | 0.31 |
ENSMUST00000182476.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr5_-_87092546 | 0.31 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr7_+_97720839 | 0.31 |
ENSMUST00000070021.1
|
Gm9990
|
predicted gene 9990 |
| chr11_+_69991633 | 0.31 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr2_+_34874396 | 0.30 |
ENSMUST00000113068.2
ENSMUST00000047447.8 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr7_-_127890918 | 0.29 |
ENSMUST00000121394.1
|
Prss53
|
protease, serine, 53 |
| chr4_-_115647709 | 0.29 |
ENSMUST00000102707.3
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr6_+_30512283 | 0.29 |
ENSMUST00000031798.7
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chrM_+_8600 | 0.28 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr2_-_67194695 | 0.27 |
ENSMUST00000147939.1
|
Gm13598
|
predicted gene 13598 |
| chr2_+_180071793 | 0.26 |
ENSMUST00000108901.1
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr5_+_114853660 | 0.26 |
ENSMUST00000031547.5
|
4930519G04Rik
|
RIKEN cDNA 4930519G04 gene |
| chr13_-_74350206 | 0.25 |
ENSMUST00000022062.7
|
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr5_-_136615328 | 0.25 |
ENSMUST00000053319.2
|
4731417B20Rik
|
RIKEN cDNA 4731417B20 gene |
| chr10_+_77864623 | 0.24 |
ENSMUST00000092366.2
|
Tspear
|
thrombospondin type laminin G domain and EAR repeats |
| chr8_+_105701142 | 0.24 |
ENSMUST00000098444.2
|
Pard6a
|
par-6 (partitioning defective 6,) homolog alpha (C. elegans) |
| chr2_+_34874486 | 0.24 |
ENSMUST00000028228.3
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr4_+_135963720 | 0.23 |
ENSMUST00000102541.3
|
Gale
|
galactose-4-epimerase, UDP |
| chr8_+_21427456 | 0.23 |
ENSMUST00000095424.4
|
Gm21498
|
predicted gene, 21498 |
| chr6_-_59024470 | 0.23 |
ENSMUST00000089860.5
|
Fam13a
|
family with sequence similarity 13, member A |
| chr13_+_4191163 | 0.23 |
ENSMUST00000021634.2
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr11_+_9048575 | 0.23 |
ENSMUST00000043285.4
|
Gm11992
|
predicted gene 11992 |
| chr6_+_141524379 | 0.21 |
ENSMUST00000032362.9
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr4_+_139622842 | 0.21 |
ENSMUST00000039818.9
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr13_-_4279420 | 0.21 |
ENSMUST00000021632.3
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr4_-_119383732 | 0.20 |
ENSMUST00000044781.2
ENSMUST00000084307.3 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr13_+_55399648 | 0.20 |
ENSMUST00000057167.7
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr2_+_164562579 | 0.20 |
ENSMUST00000017867.3
ENSMUST00000109344.2 ENSMUST00000109345.2 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr5_-_66080971 | 0.19 |
ENSMUST00000127275.1
ENSMUST00000113724.1 |
Rbm47
|
RNA binding motif protein 47 |
| chr10_+_4611971 | 0.19 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr3_+_87435820 | 0.19 |
ENSMUST00000178261.1
ENSMUST00000049926.8 ENSMUST00000166297.1 |
Fcrl5
|
Fc receptor-like 5 |
| chr10_+_120208739 | 0.19 |
ENSMUST00000020446.4
ENSMUST00000134797.1 |
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr19_-_16873830 | 0.19 |
ENSMUST00000072915.2
|
Foxb2
|
forkhead box B2 |
| chr7_+_44216456 | 0.19 |
ENSMUST00000074359.2
|
Klk1b5
|
kallikrein 1-related peptidase b5 |
| chrX_-_134161928 | 0.18 |
ENSMUST00000033611.4
|
Xkrx
|
X Kell blood group precursor related X linked |
| chr13_+_74406387 | 0.18 |
ENSMUST00000090860.6
|
Gm10116
|
predicted pseudogene 10116 |
| chr14_-_78088994 | 0.18 |
ENSMUST00000118785.2
ENSMUST00000066437.4 |
Fam216b
|
family with sequence similarity 216, member B |
| chr10_+_43901782 | 0.18 |
ENSMUST00000054418.5
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr12_-_114406773 | 0.17 |
ENSMUST00000177949.1
|
Ighv6-4
|
immunoglobulin heavy variable V6-4 |
| chr7_+_141949982 | 0.17 |
ENSMUST00000105989.2
ENSMUST00000075528.5 ENSMUST00000174499.1 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr7_+_44849949 | 0.17 |
ENSMUST00000141311.1
ENSMUST00000107880.1 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr6_-_144209448 | 0.17 |
ENSMUST00000077160.5
|
Sox5
|
SRY-box containing gene 5 |
| chr8_-_22653406 | 0.17 |
ENSMUST00000033938.5
|
Polb
|
polymerase (DNA directed), beta |
| chr6_-_144209471 | 0.17 |
ENSMUST00000038815.7
|
Sox5
|
SRY-box containing gene 5 |
| chr14_-_51256112 | 0.17 |
ENSMUST00000061936.6
|
Ear11
|
eosinophil-associated, ribonuclease A family, member 11 |
| chr11_-_98149551 | 0.16 |
ENSMUST00000103143.3
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr12_-_75177325 | 0.16 |
ENSMUST00000042299.2
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr1_+_74601548 | 0.16 |
ENSMUST00000087186.4
|
Stk36
|
serine/threonine kinase 36 |
| chr15_-_36140393 | 0.16 |
ENSMUST00000172831.1
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr6_-_59024340 | 0.15 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr19_-_6942406 | 0.15 |
ENSMUST00000099782.3
|
Gpr137
|
G protein-coupled receptor 137 |
| chr13_-_40733768 | 0.15 |
ENSMUST00000110193.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr4_-_116555896 | 0.15 |
ENSMUST00000069674.5
ENSMUST00000106478.2 |
Tmem69
|
transmembrane protein 69 |
| chr16_+_17070281 | 0.15 |
ENSMUST00000090199.3
|
Ypel1
|
yippee-like 1 (Drosophila) |
| chr7_+_105404568 | 0.15 |
ENSMUST00000033187.4
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr1_-_179546261 | 0.15 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr2_-_129048172 | 0.15 |
ENSMUST00000145798.2
|
Gm14025
|
predicted gene 14025 |
| chr1_+_107361929 | 0.14 |
ENSMUST00000027566.2
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr4_+_134496996 | 0.14 |
ENSMUST00000095074.3
|
Paqr7
|
progestin and adipoQ receptor family member VII |
| chr1_-_75046639 | 0.14 |
ENSMUST00000152855.1
|
Nhej1
|
nonhomologous end-joining factor 1 |
| chr2_+_74695601 | 0.14 |
ENSMUST00000152027.1
|
Gm14396
|
predicted gene 14396 |
| chr4_+_135963742 | 0.14 |
ENSMUST00000149636.1
ENSMUST00000143304.1 |
Gale
|
galactose-4-epimerase, UDP |
| chr9_-_16378231 | 0.14 |
ENSMUST00000082170.5
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila) |
| chr8_+_3621529 | 0.14 |
ENSMUST00000156380.2
|
Pet100
|
PET100 homolog (S. cerevisiae) |
| chr12_-_70231414 | 0.14 |
ENSMUST00000161083.1
|
Pygl
|
liver glycogen phosphorylase |
| chr15_+_78935177 | 0.13 |
ENSMUST00000145157.1
ENSMUST00000123013.1 |
Nol12
|
nucleolar protein 12 |
| chr14_+_77036746 | 0.13 |
ENSMUST00000048208.3
ENSMUST00000095625.4 |
Ccdc122
|
coiled-coil domain containing 122 |
| chr4_-_43771009 | 0.13 |
ENSMUST00000053931.1
|
Olfr159
|
olfactory receptor 159 |
| chr9_+_7347374 | 0.13 |
ENSMUST00000065079.5
ENSMUST00000005950.5 |
Mmp12
|
matrix metallopeptidase 12 |
| chr5_+_124540695 | 0.13 |
ENSMUST00000060226.4
|
Tmed2
|
transmembrane emp24 domain trafficking protein 2 |
| chr11_-_70239794 | 0.13 |
ENSMUST00000040428.3
|
Rnasek
|
ribonuclease, RNase K |
| chrY_+_5658985 | 0.13 |
ENSMUST00000179901.1
|
Gm21854
|
predicted gene, 21854 |
| chr2_+_103411682 | 0.13 |
ENSMUST00000164172.1
|
Elf5
|
E74-like factor 5 |
| chr11_-_99993992 | 0.13 |
ENSMUST00000105049.1
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr9_+_45906513 | 0.13 |
ENSMUST00000039059.6
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr6_-_144209558 | 0.13 |
ENSMUST00000111749.1
ENSMUST00000170367.2 |
Sox5
|
SRY-box containing gene 5 |
| chr11_-_100759740 | 0.13 |
ENSMUST00000107361.2
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr5_-_112589990 | 0.12 |
ENSMUST00000183186.1
ENSMUST00000182949.1 |
1700034G24Rik
|
RIKEN cDNA 1700034G24 gene |
| chr2_+_177074320 | 0.12 |
ENSMUST00000108970.2
|
Gm14401
|
predicted gene 14401 |
| chr6_-_86733268 | 0.12 |
ENSMUST00000001185.7
|
Gmcl1
|
germ cell-less homolog 1 (Drosophila) |
| chrX_+_139563316 | 0.12 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr12_+_70974621 | 0.12 |
ENSMUST00000160027.1
ENSMUST00000160864.1 |
Psma3
|
proteasome (prosome, macropain) subunit, alpha type 3 |
| chr8_+_9864251 | 0.12 |
ENSMUST00000088080.2
|
Gm10217
|
predicted gene 10217 |
| chrY_+_13495642 | 0.12 |
ENSMUST00000178940.1
|
Gm20773
|
predicted gene, 20773 |
| chr14_+_30549131 | 0.12 |
ENSMUST00000022529.6
|
Tkt
|
transketolase |
| chr3_-_98762404 | 0.12 |
ENSMUST00000146196.1
ENSMUST00000148578.1 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr14_+_120275669 | 0.12 |
ENSMUST00000088419.6
ENSMUST00000167459.1 |
Mbnl2
|
muscleblind-like 2 |
| chr14_+_64104286 | 0.12 |
ENSMUST00000022537.5
|
Prss52
|
protease, serine, 52 |
| chr9_+_75347755 | 0.12 |
ENSMUST00000034709.5
|
Bcl2l10
|
Bcl2-like 10 |
| chr11_-_105456674 | 0.11 |
ENSMUST00000049995.3
ENSMUST00000100332.3 |
March10
|
membrane-associated ring finger (C3HC4) 10 |
| chr1_+_44119952 | 0.11 |
ENSMUST00000114709.2
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr6_-_42472200 | 0.11 |
ENSMUST00000170504.1
|
Olfr457
|
olfactory receptor 457 |
| chr10_+_78425670 | 0.11 |
ENSMUST00000005185.6
|
Cstb
|
cystatin B |
| chr10_+_82058417 | 0.11 |
ENSMUST00000105313.1
|
Zfp873
|
zinc finger protein 873 |
| chr15_+_85510812 | 0.11 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr4_+_134496966 | 0.11 |
ENSMUST00000134506.1
ENSMUST00000136171.1 ENSMUST00000125330.1 |
Paqr7
|
progestin and adipoQ receptor family member VII |
| chr17_-_31144271 | 0.11 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr2_+_172248492 | 0.11 |
ENSMUST00000038532.1
|
Mc3r
|
melanocortin 3 receptor |
| chr4_-_124610803 | 0.10 |
ENSMUST00000053604.2
|
3100002H09Rik
|
RIKEN cDNA 3100002H09 gene |
| chr19_+_18631927 | 0.10 |
ENSMUST00000159572.1
ENSMUST00000042392.7 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr7_+_141949688 | 0.10 |
ENSMUST00000018971.8
ENSMUST00000078200.5 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr7_+_128203598 | 0.10 |
ENSMUST00000177383.1
|
Itgad
|
integrin, alpha D |
| chr7_+_141949846 | 0.10 |
ENSMUST00000172652.1
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr7_-_57509995 | 0.10 |
ENSMUST00000068456.6
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
| chr14_-_20480106 | 0.10 |
ENSMUST00000065504.9
ENSMUST00000100844.4 |
Anxa7
|
annexin A7 |
| chr8_+_45975514 | 0.10 |
ENSMUST00000034051.6
|
Ufsp2
|
UFM1-specific peptidase 2 |
| chr2_-_19553908 | 0.10 |
ENSMUST00000062060.4
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr7_+_96951505 | 0.09 |
ENSMUST00000044466.5
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chrY_+_38605676 | 0.09 |
ENSMUST00000180228.1
|
Gm21765
|
predicted gene, 21765 |
| chrY_-_87447885 | 0.09 |
ENSMUST00000178784.1
|
Gm21813
|
predicted gene, 21813 |
| chr14_+_66937365 | 0.09 |
ENSMUST00000052898.3
|
4930578I07Rik
|
RIKEN cDNA 4930578I07 gene |
| chr2_-_177925604 | 0.09 |
ENSMUST00000108934.2
ENSMUST00000081529.4 |
C330013J21Rik
|
RIKEN cDNA C330013J21 gene |
| chr3_-_19628669 | 0.09 |
ENSMUST00000119133.1
|
1700064H15Rik
|
RIKEN cDNA 1700064H15 gene |
| chr19_-_41206774 | 0.09 |
ENSMUST00000025986.7
ENSMUST00000169941.1 |
Tll2
|
tolloid-like 2 |
| chr5_-_49285653 | 0.09 |
ENSMUST00000175660.1
|
Kcnip4
|
Kv channel interacting protein 4 |
| chrY_+_66123648 | 0.09 |
ENSMUST00000180028.1
|
Gm21837
|
predicted gene, 21837 |
| chrY_-_53486001 | 0.09 |
ENSMUST00000178580.1
|
Gm21906
|
predicted gene, 21906 |
| chrY_-_82163360 | 0.09 |
ENSMUST00000179406.1
|
Gm21825
|
predicted gene, 21825 |
| chr9_-_51328898 | 0.09 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chrY_-_10750826 | 0.09 |
ENSMUST00000177818.1
|
Gm20777
|
predicted gene, 20777 |
| chr19_-_18631754 | 0.09 |
ENSMUST00000025631.6
|
Ostf1
|
osteoclast stimulating factor 1 |
| chr2_+_122028544 | 0.09 |
ENSMUST00000028668.7
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chrX_-_38043105 | 0.08 |
ENSMUST00000089060.3
|
4930430D24Rik
|
RIKEN cDNA 4930430D24 gene |
| chr2_-_69885557 | 0.08 |
ENSMUST00000060447.6
|
Mettl5
|
methyltransferase like 5 |
| chr10_-_57532489 | 0.08 |
ENSMUST00000020027.4
|
Serinc1
|
serine incorporator 1 |
| chrY_-_84629416 | 0.08 |
ENSMUST00000179495.1
|
Gm21824
|
predicted gene, 21824 |
| chrX_+_8413305 | 0.08 |
ENSMUST00000037297.3
|
Ssxb1
|
synovial sarcoma, X member B, breakpoint 1 |
| chrX_-_89409689 | 0.08 |
ENSMUST00000113959.1
ENSMUST00000113960.2 |
Pet2
|
plasmacytoma expressed transcript 2 |
| chrY_-_51021720 | 0.08 |
ENSMUST00000178596.1
|
Gm21849
|
predicted gene, 21849 |
| chrY_-_52328958 | 0.08 |
ENSMUST00000180160.1
|
Gm21899
|
predicted gene, 21899 |
| chrY_-_64925708 | 0.08 |
ENSMUST00000179378.1
|
Gm21826
|
predicted gene, 21826 |
| chrY_-_72104014 | 0.08 |
ENSMUST00000179902.1
|
Gm21880
|
predicted gene, 21880 |
| chr9_-_55283582 | 0.08 |
ENSMUST00000114306.1
ENSMUST00000164721.1 |
Nrg4
|
neuregulin 4 |
| chr8_+_28593505 | 0.08 |
ENSMUST00000180993.1
ENSMUST00000181267.1 |
Gm26795
|
predicted gene, 26795 |
| chrX_-_38018815 | 0.08 |
ENSMUST00000184432.1
|
4930525M21RIK
|
B cell translocation gene 1, anti-proliferative-like |
| chr15_-_78718113 | 0.08 |
ENSMUST00000088592.4
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chrY_+_40682046 | 0.08 |
ENSMUST00000178175.1
|
Gm20793
|
predicted gene, 20793 |
| chrY_+_40806531 | 0.08 |
ENSMUST00000178528.1
|
Gm20795
|
predicted gene, 20795 |
| chrY_-_13911041 | 0.08 |
ENSMUST00000179339.1
|
Gm20836
|
predicted gene, 20836 |
| chrY_-_18623762 | 0.08 |
ENSMUST00000178373.1
|
Gm21660
|
predicted gene, 21660 |
| chrX_+_107667956 | 0.08 |
ENSMUST00000168174.2
|
Tbx22
|
T-box 22 |
| chrY_-_81869220 | 0.08 |
ENSMUST00000179012.1
|
Gm21919
|
predicted gene, 21919 |
| chrY_-_10534428 | 0.08 |
ENSMUST00000179665.1
|
Gm20737
|
predicted gene, 20737 |
| chrY_+_18090734 | 0.08 |
ENSMUST00000180056.1
|
Ssty1
|
spermiogenesis specific transcript on the Y 1 |
| chrY_+_21096898 | 0.08 |
ENSMUST00000180223.1
|
Gm20910
|
predicted gene, 20910 |
| chrY_-_41376429 | 0.08 |
ENSMUST00000178470.1
|
Gm20827
|
predicted gene, 20827 |
| chrY_+_18208481 | 0.08 |
ENSMUST00000177902.1
|
Gm20851
|
predicted gene, 20851 |
| chr8_+_70131327 | 0.08 |
ENSMUST00000095273.5
|
Nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chrX_+_53434918 | 0.08 |
ENSMUST00000074232.6
|
Etd
|
embryonic testis differentiation |
| chrY_+_35503137 | 0.08 |
ENSMUST00000180125.1
|
Gm21795
|
predicted gene, 21795 |
| chrY_-_71364937 | 0.08 |
ENSMUST00000177885.1
|
Gm21841
|
predicted gene, 21841 |
| chrY_-_83994275 | 0.08 |
ENSMUST00000177980.1
|
Gm21717
|
predicted gene, 21717 |
| chrY_+_37818134 | 0.08 |
ENSMUST00000179746.1
|
Gm21878
|
predicted gene, 21878 |
| chr4_-_88691268 | 0.08 |
ENSMUST00000105146.2
|
Ifnab
|
interferon alpha B |
| chr2_-_89752826 | 0.08 |
ENSMUST00000099765.1
|
Olfr1253
|
olfactory receptor 1253 |
| chr6_-_87851011 | 0.08 |
ENSMUST00000113617.1
|
Cnbp
|
cellular nucleic acid binding protein |
| chr9_-_109495875 | 0.07 |
ENSMUST00000076617.4
|
Fbxw19
|
F-box and WD-40 domain protein 19 |
| chrY_+_23240330 | 0.07 |
ENSMUST00000178468.1
|
Gm21907
|
predicted gene, 21907 |
| chrY_-_72450406 | 0.07 |
ENSMUST00000179653.1
|
Gm21757
|
predicted gene, 21757 |
| chrY_-_73383907 | 0.07 |
ENSMUST00000180192.1
|
Gm21855
|
predicted gene, 21855 |
| chrY_-_86500240 | 0.07 |
ENSMUST00000179810.1
|
Gm21796
|
predicted gene, 21796 |
| chrY_+_23802239 | 0.07 |
ENSMUST00000180287.1
|
Gm21768
|
predicted gene, 21768 |
| chrY_+_36025254 | 0.07 |
ENSMUST00000178190.1
|
Gm21898
|
predicted gene, 21898 |
| chrY_-_52675541 | 0.07 |
ENSMUST00000178917.1
|
Gm21840
|
predicted gene, 21840 |
| chrY_-_65273203 | 0.07 |
ENSMUST00000177863.1
|
Gm21848
|
predicted gene, 21848 |
| chr2_+_83644435 | 0.07 |
ENSMUST00000081591.6
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr12_+_52097737 | 0.07 |
ENSMUST00000040090.9
|
Nubpl
|
nucleotide binding protein-like |
| chrY_-_88261004 | 0.07 |
ENSMUST00000177900.1
|
Gm21777
|
predicted gene, 21777 |
| chr6_-_42597372 | 0.07 |
ENSMUST00000069023.3
|
Fam115e
|
family with sequence similarity 115, member E |
| chr6_+_114403825 | 0.07 |
ENSMUST00000161220.1
|
Hrh1
|
histamine receptor H1 |
| chrY_-_81352055 | 0.07 |
ENSMUST00000179831.1
|
Gm21829
|
predicted gene, 21829 |
| chr9_-_45906837 | 0.07 |
ENSMUST00000161203.1
ENSMUST00000058720.5 ENSMUST00000160699.1 |
Rnf214
|
ring finger protein 214 |
| chr10_-_63927434 | 0.07 |
ENSMUST00000079279.3
|
Gm10118
|
predicted gene 10118 |
| chr11_-_94507337 | 0.07 |
ENSMUST00000040692.8
|
Mycbpap
|
MYCBP associated protein |
| chr7_-_119720742 | 0.07 |
ENSMUST00000033236.7
|
Thumpd1
|
THUMP domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Osr2_Osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.1 | 1.0 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.5 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.3 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.4 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.1 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.4 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.2 | GO:0021623 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.4 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0051381 | histamine binding(GO:0051381) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |