Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
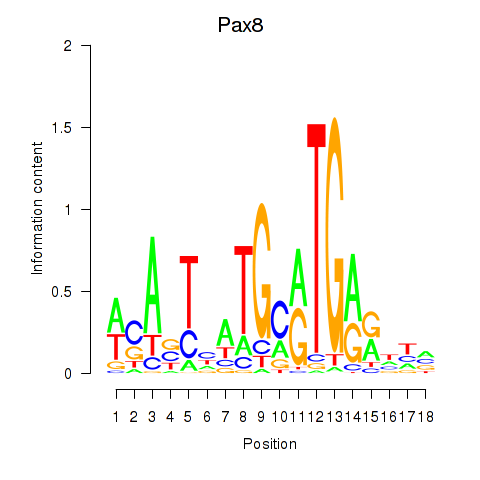
Results for Pax8
Z-value: 0.39
Transcription factors associated with Pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax8
|
ENSMUSG00000026976.9 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax8 | mm10_v2_chr2_-_24475596_24475605 | -0.37 | 2.4e-02 | Click! |
Activity profile of Pax8 motif
Sorted Z-values of Pax8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_52863299 | 0.58 |
ENSMUST00000103616.3
|
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr13_-_62607499 | 0.56 |
ENSMUST00000091563.4
|
6720489N17Rik
|
RIKEN cDNA 6720489N17 gene |
| chr5_-_87591582 | 0.55 |
ENSMUST00000031201.7
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr1_-_139608282 | 0.53 |
ENSMUST00000170441.2
|
Cfhr3
|
complement factor H-related 3 |
| chr14_+_53676141 | 0.50 |
ENSMUST00000103662.4
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr7_+_112742025 | 0.48 |
ENSMUST00000164363.1
|
Tead1
|
TEA domain family member 1 |
| chr16_+_34784917 | 0.27 |
ENSMUST00000023538.8
|
Mylk
|
myosin, light polypeptide kinase |
| chr2_+_177508570 | 0.23 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr11_-_49114874 | 0.21 |
ENSMUST00000109201.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr6_+_127072902 | 0.21 |
ENSMUST00000000186.6
|
Fgf23
|
fibroblast growth factor 23 |
| chr9_+_109052828 | 0.19 |
ENSMUST00000124432.1
|
Shisa5
|
shisa homolog 5 (Xenopus laevis) |
| chrX_+_82948861 | 0.19 |
ENSMUST00000114000.1
|
Dmd
|
dystrophin, muscular dystrophy |
| chrX_-_7572843 | 0.19 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr2_-_150255591 | 0.19 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr13_-_62520451 | 0.18 |
ENSMUST00000082203.6
ENSMUST00000101547.4 |
Zfp934
|
zinc finger protein 934 |
| chr13_-_66852017 | 0.16 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr7_-_30861470 | 0.15 |
ENSMUST00000052700.3
|
Ffar1
|
free fatty acid receptor 1 |
| chr9_-_103230262 | 0.14 |
ENSMUST00000165296.1
ENSMUST00000112645.1 ENSMUST00000166836.1 |
Trf
Gm20425
|
transferrin predicted gene 20425 |
| chr9_-_103230415 | 0.14 |
ENSMUST00000035158.9
|
Trf
|
transferrin |
| chr7_-_29906087 | 0.14 |
ENSMUST00000053521.8
|
Zfp27
|
zinc finger protein 27 |
| chr12_+_71048338 | 0.13 |
ENSMUST00000135709.1
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_114768767 | 0.10 |
ENSMUST00000151464.1
|
Insc
|
inscuteable homolog (Drosophila) |
| chr10_+_18845071 | 0.10 |
ENSMUST00000019998.7
|
Perp
|
PERP, TP53 apoptosis effector |
| chr5_+_91074611 | 0.10 |
ENSMUST00000031324.4
|
Ereg
|
epiregulin |
| chr1_+_174050375 | 0.10 |
ENSMUST00000062665.3
|
Olfr432
|
olfactory receptor 432 |
| chr15_+_54410755 | 0.09 |
ENSMUST00000036737.3
|
Colec10
|
collectin sub-family member 10 |
| chr11_+_78499087 | 0.09 |
ENSMUST00000017488.4
|
Vtn
|
vitronectin |
| chr4_+_103313806 | 0.09 |
ENSMUST00000035780.3
|
Oma1
|
OMA1 homolog, zinc metallopeptidase (S. cerevisiae) |
| chrX_-_105391770 | 0.09 |
ENSMUST00000136406.1
ENSMUST00000150604.1 |
5330434G04Rik
|
RIKEN cDNA 5330434G04 gene |
| chr7_+_107595051 | 0.08 |
ENSMUST00000040056.7
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chrX_-_105391726 | 0.08 |
ENSMUST00000142109.1
|
5330434G04Rik
|
RIKEN cDNA 5330434G04 gene |
| chr1_+_65186727 | 0.08 |
ENSMUST00000097707.4
ENSMUST00000081154.7 |
Pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr13_-_62777089 | 0.07 |
ENSMUST00000167516.2
|
Gm5141
|
predicted gene 5141 |
| chr7_-_42578588 | 0.07 |
ENSMUST00000179470.1
|
Gm21028
|
predicted gene, 21028 |
| chr12_-_20900867 | 0.07 |
ENSMUST00000079237.5
|
Zfp125
|
zinc finger protein 125 |
| chr7_+_114768736 | 0.07 |
ENSMUST00000117543.1
|
Insc
|
inscuteable homolog (Drosophila) |
| chr1_-_92518515 | 0.06 |
ENSMUST00000062353.4
|
Olfr1414
|
olfactory receptor 1414 |
| chr6_+_139736895 | 0.06 |
ENSMUST00000111868.3
|
Pik3c2g
|
phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide |
| chr15_-_101869695 | 0.05 |
ENSMUST00000087996.5
|
Krt77
|
keratin 77 |
| chr12_+_84996487 | 0.04 |
ENSMUST00000101202.3
|
Ylpm1
|
YLP motif containing 1 |
| chr8_-_64849818 | 0.04 |
ENSMUST00000034017.7
|
Klhl2
|
kelch-like 2, Mayven |
| chr7_-_141100526 | 0.03 |
ENSMUST00000097958.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr16_-_19408078 | 0.03 |
ENSMUST00000052516.4
|
Olfr165
|
olfactory receptor 165 |
| chr5_+_100196611 | 0.03 |
ENSMUST00000066813.1
|
Gm9932
|
predicted gene 9932 |
| chr1_+_6487231 | 0.03 |
ENSMUST00000140079.1
ENSMUST00000131494.1 |
St18
|
suppression of tumorigenicity 18 |
| chr7_+_12897800 | 0.02 |
ENSMUST00000055528.4
ENSMUST00000117189.1 ENSMUST00000120809.1 ENSMUST00000119989.1 |
Zscan22
|
zinc finger and SCAN domain containing 22 |
| chr12_-_28582515 | 0.02 |
ENSMUST00000110917.1
ENSMUST00000020965.7 |
Allc
|
allantoicase |
| chr19_+_46075842 | 0.01 |
ENSMUST00000165017.1
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr9_+_32116040 | 0.00 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr17_+_21691860 | 0.00 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.5 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |