Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
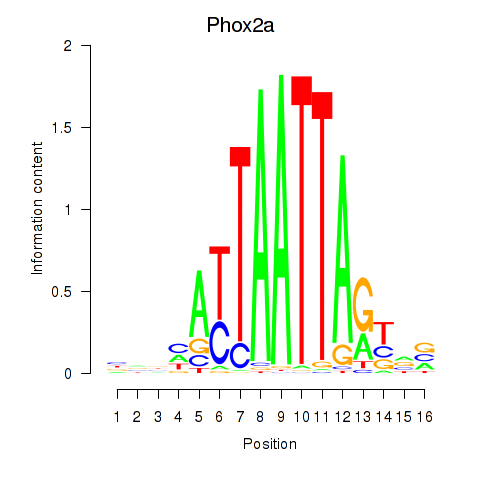
Results for Phox2a
Z-value: 1.02
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSMUSG00000007946.9 | paired-like homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2a | mm10_v2_chr7_+_101818306_101818321 | 0.65 | 2.0e-05 | Click! |
Activity profile of Phox2a motif
Sorted Z-values of Phox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_168767136 | 6.66 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr2_-_168767029 | 6.12 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr16_-_42340595 | 5.53 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr14_+_27000362 | 4.00 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chrX_-_139871637 | 3.99 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chr6_-_30958990 | 3.91 |
ENSMUST00000101589.3
|
Klf14
|
Kruppel-like factor 14 |
| chr1_-_89933290 | 3.40 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr9_+_119063429 | 3.00 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chrX_+_9885622 | 2.19 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr15_-_34356421 | 1.59 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr19_-_55241236 | 1.49 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr19_-_47692042 | 1.46 |
ENSMUST00000026045.7
ENSMUST00000086923.5 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr1_+_172555932 | 1.35 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr1_-_172027269 | 1.33 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr11_-_99244058 | 1.09 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr5_+_13398688 | 1.01 |
ENSMUST00000125629.1
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_+_36230426 | 0.91 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr6_+_92816460 | 0.80 |
ENSMUST00000057977.3
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr5_-_118244861 | 0.75 |
ENSMUST00000117177.1
ENSMUST00000133372.1 ENSMUST00000154786.1 ENSMUST00000121369.1 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr2_-_164585447 | 0.56 |
ENSMUST00000109342.1
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr13_-_102906046 | 0.54 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_-_72967854 | 0.41 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr16_-_29544852 | 0.36 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr8_+_107031218 | 0.30 |
ENSMUST00000034388.9
|
Vps4a
|
vacuolar protein sorting 4a (yeast) |
| chr18_+_55057557 | 0.27 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
| chr3_+_66219909 | 0.23 |
ENSMUST00000029421.5
|
Ptx3
|
pentraxin related gene |
| chr4_-_14621805 | 0.21 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_+_134930898 | 0.09 |
ENSMUST00000030622.2
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Phox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 5.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 3.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.4 | 1.3 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 | 3.4 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.4 | 4.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.0 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 0.9 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.3 | GO:1903774 | mitotic cytokinesis checkpoint(GO:0044878) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.6 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.1 | 0.2 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 5.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 12.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 7.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) chemorepellent activity(GO:0045499) |
| 0.1 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 2.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 4.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 12.4 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 12.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |