Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
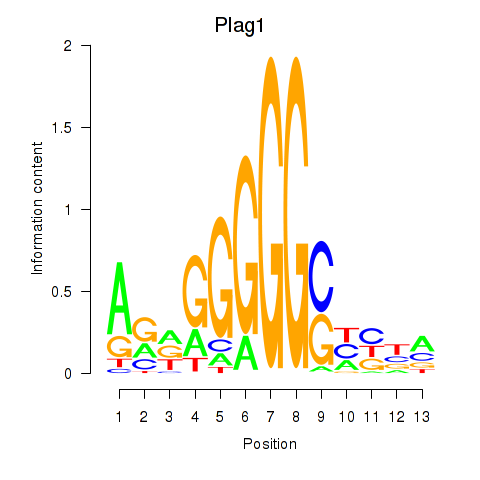
Results for Plag1
Z-value: 2.75
Transcription factors associated with Plag1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plag1
|
ENSMUSG00000003282.3 | pleiomorphic adenoma gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plag1 | mm10_v2_chr4_-_3938354_3938401 | 0.66 | 1.2e-05 | Click! |
Activity profile of Plag1 motif
Sorted Z-values of Plag1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_80000292 | 10.52 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr7_-_142657466 | 10.45 |
ENSMUST00000097936.2
ENSMUST00000000033.5 |
Igf2
|
insulin-like growth factor 2 |
| chr11_+_11684967 | 10.22 |
ENSMUST00000126058.1
ENSMUST00000141436.1 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr7_-_142659482 | 9.85 |
ENSMUST00000121128.1
|
Igf2
|
insulin-like growth factor 2 |
| chr11_+_74619594 | 9.05 |
ENSMUST00000100866.2
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene |
| chr16_-_18622403 | 8.75 |
ENSMUST00000167388.1
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr18_-_58209926 | 8.63 |
ENSMUST00000025497.6
|
Fbn2
|
fibrillin 2 |
| chr10_+_75948292 | 8.28 |
ENSMUST00000000926.2
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chrX_+_8271133 | 8.02 |
ENSMUST00000127103.1
ENSMUST00000115591.1 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr11_-_102082464 | 7.94 |
ENSMUST00000100398.4
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr5_+_33995984 | 6.80 |
ENSMUST00000056355.8
|
Nat8l
|
N-acetyltransferase 8-like |
| chr11_-_87875524 | 6.78 |
ENSMUST00000049768.3
|
Epx
|
eosinophil peroxidase |
| chr1_-_45503282 | 6.69 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr9_-_44288535 | 6.60 |
ENSMUST00000161354.1
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr1_-_75133866 | 6.58 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chrX_-_102252154 | 6.21 |
ENSMUST00000050551.3
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr15_+_89334398 | 6.12 |
ENSMUST00000023282.2
|
Miox
|
myo-inositol oxygenase |
| chr9_-_44288332 | 6.07 |
ENSMUST00000161408.1
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr8_-_10928449 | 5.67 |
ENSMUST00000040608.3
|
3930402G23Rik
|
RIKEN cDNA 3930402G23 gene |
| chr19_+_8591254 | 5.62 |
ENSMUST00000010251.3
ENSMUST00000170817.1 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr2_+_163054682 | 5.62 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr6_+_90619241 | 5.57 |
ENSMUST00000032177.8
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr15_+_75156693 | 5.56 |
ENSMUST00000023246.3
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr7_-_143460989 | 5.36 |
ENSMUST00000167912.1
ENSMUST00000037287.6 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr4_+_115059507 | 5.28 |
ENSMUST00000162489.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr17_-_87797994 | 5.27 |
ENSMUST00000055221.7
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr12_-_76709997 | 4.99 |
ENSMUST00000166101.1
|
Sptb
|
spectrin beta, erythrocytic |
| chr11_+_115877497 | 4.73 |
ENSMUST00000144032.1
|
Myo15b
|
myosin XVB |
| chr1_-_189688074 | 4.64 |
ENSMUST00000171929.1
ENSMUST00000165962.1 |
Cenpf
|
centromere protein F |
| chr12_-_4841583 | 4.61 |
ENSMUST00000020964.5
|
Fkbp1b
|
FK506 binding protein 1b |
| chr17_+_36869567 | 4.60 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr7_-_4752972 | 4.53 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr15_+_79892436 | 4.52 |
ENSMUST00000175752.1
ENSMUST00000176325.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr8_-_71725696 | 4.47 |
ENSMUST00000153800.1
ENSMUST00000146100.1 |
Fcho1
|
FCH domain only 1 |
| chr4_+_115057683 | 4.46 |
ENSMUST00000161601.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr3_+_10012548 | 4.43 |
ENSMUST00000029046.8
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr4_-_132533488 | 4.35 |
ENSMUST00000152993.1
ENSMUST00000067496.6 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr7_+_45335256 | 4.34 |
ENSMUST00000085351.4
|
Hrc
|
histidine rich calcium binding protein |
| chr11_+_24080664 | 4.33 |
ENSMUST00000118955.1
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr4_+_130308595 | 4.29 |
ENSMUST00000070532.7
|
Fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr7_+_79660196 | 4.28 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr1_+_75382114 | 4.23 |
ENSMUST00000113590.1
ENSMUST00000148515.1 |
Speg
|
SPEG complex locus |
| chr7_+_78914216 | 4.19 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr19_-_34166037 | 4.19 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr15_+_79892397 | 4.11 |
ENSMUST00000175714.1
ENSMUST00000109620.3 ENSMUST00000165537.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr17_+_29318850 | 3.99 |
ENSMUST00000114701.2
|
Pi16
|
peptidase inhibitor 16 |
| chr2_-_91931774 | 3.93 |
ENSMUST00000069423.6
|
Mdk
|
midkine |
| chr10_-_102490418 | 3.82 |
ENSMUST00000020040.3
|
Nts
|
neurotensin |
| chr1_+_134182404 | 3.80 |
ENSMUST00000153856.1
ENSMUST00000082060.3 ENSMUST00000133701.1 ENSMUST00000132873.1 |
Chi3l1
|
chitinase 3-like 1 |
| chrX_-_93832106 | 3.78 |
ENSMUST00000045748.6
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr3_+_108364882 | 3.78 |
ENSMUST00000090563.5
|
Mybphl
|
myosin binding protein H-like |
| chr14_-_60087347 | 3.78 |
ENSMUST00000149414.1
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr11_+_78343475 | 3.76 |
ENSMUST00000002127.7
ENSMUST00000108295.1 |
Unc119
|
unc-119 homolog (C. elegans) |
| chr11_-_55185029 | 3.76 |
ENSMUST00000039305.5
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr2_+_84840612 | 3.74 |
ENSMUST00000111625.1
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr10_+_75566257 | 3.73 |
ENSMUST00000129232.1
ENSMUST00000143792.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr5_-_145584723 | 3.71 |
ENSMUST00000075837.6
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr9_+_62858085 | 3.69 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chr4_-_152477433 | 3.64 |
ENSMUST00000159186.1
ENSMUST00000162017.1 ENSMUST00000030768.2 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr7_-_126369543 | 3.60 |
ENSMUST00000032997.6
|
Lat
|
linker for activation of T cells |
| chr15_-_76669811 | 3.56 |
ENSMUST00000037824.4
|
Foxh1
|
forkhead box H1 |
| chr17_-_56830916 | 3.55 |
ENSMUST00000002444.7
ENSMUST00000086801.5 |
Rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr15_-_103251465 | 3.50 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_-_32353247 | 3.50 |
ENSMUST00000078352.5
ENSMUST00000113352.2 ENSMUST00000113365.1 |
Dnm1
|
dynamin 1 |
| chr5_-_37717122 | 3.49 |
ENSMUST00000094836.4
|
Stk32b
|
serine/threonine kinase 32B |
| chr17_-_46629420 | 3.47 |
ENSMUST00000044442.8
|
Ptk7
|
PTK7 protein tyrosine kinase 7 |
| chr9_-_44288131 | 3.47 |
ENSMUST00000160384.1
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr3_+_88532314 | 3.43 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr11_+_7197780 | 3.42 |
ENSMUST00000020704.7
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr17_+_43568641 | 3.39 |
ENSMUST00000169694.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr10_+_13090788 | 3.32 |
ENSMUST00000121646.1
ENSMUST00000121325.1 ENSMUST00000121766.1 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr17_+_43568475 | 3.27 |
ENSMUST00000167418.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr6_-_126939524 | 3.27 |
ENSMUST00000144954.1
ENSMUST00000112221.1 ENSMUST00000112220.1 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr7_+_43351378 | 3.27 |
ENSMUST00000012798.7
ENSMUST00000122423.1 ENSMUST00000121494.1 |
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr2_+_157560078 | 3.25 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr17_+_48299952 | 3.23 |
ENSMUST00000170941.1
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr16_+_4594683 | 3.16 |
ENSMUST00000014447.6
|
Glis2
|
GLIS family zinc finger 2 |
| chr3_+_153973436 | 3.13 |
ENSMUST00000089948.5
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr15_-_103255433 | 3.13 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr6_-_125191535 | 3.09 |
ENSMUST00000043848.4
|
Ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr17_+_43568096 | 3.08 |
ENSMUST00000167214.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr7_-_120982260 | 3.08 |
ENSMUST00000033169.8
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr17_+_46650328 | 3.05 |
ENSMUST00000043464.7
|
Cul7
|
cullin 7 |
| chr7_+_110773658 | 3.05 |
ENSMUST00000143786.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chrX_-_136958000 | 3.01 |
ENSMUST00000069803.4
|
Tmsb15b2
|
thymosin beta 15b2 |
| chr7_-_45239041 | 2.99 |
ENSMUST00000131290.1
|
Cd37
|
CD37 antigen |
| chr4_-_63403330 | 2.96 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chrX_-_111463149 | 2.96 |
ENSMUST00000096348.3
ENSMUST00000113428.2 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr3_+_69004969 | 2.93 |
ENSMUST00000136502.1
ENSMUST00000107803.1 |
Smc4
|
structural maintenance of chromosomes 4 |
| chr14_-_56262233 | 2.91 |
ENSMUST00000015581.4
|
Gzmb
|
granzyme B |
| chr2_-_24763047 | 2.90 |
ENSMUST00000100348.3
ENSMUST00000041342.5 ENSMUST00000114447.1 ENSMUST00000102939.2 ENSMUST00000070864.7 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr11_+_94990996 | 2.88 |
ENSMUST00000038696.5
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr7_+_126950687 | 2.87 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr7_+_126950518 | 2.86 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr7_+_126950837 | 2.85 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr6_-_83033422 | 2.83 |
ENSMUST00000089651.5
|
Dok1
|
docking protein 1 |
| chr4_-_152131865 | 2.77 |
ENSMUST00000105653.1
|
Espn
|
espin |
| chr2_+_156840077 | 2.76 |
ENSMUST00000081335.6
ENSMUST00000073352.3 |
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr6_+_29433248 | 2.69 |
ENSMUST00000101617.2
ENSMUST00000065090.5 |
Flnc
|
filamin C, gamma |
| chr15_+_73723131 | 2.69 |
ENSMUST00000165541.1
ENSMUST00000167582.1 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr17_+_27556668 | 2.68 |
ENSMUST00000117254.1
ENSMUST00000118570.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chrX_+_50841434 | 2.66 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chr8_+_84148025 | 2.66 |
ENSMUST00000143833.1
ENSMUST00000118856.1 |
4930432K21Rik
|
RIKEN cDNA 4930432K21 gene |
| chr10_-_76961788 | 2.65 |
ENSMUST00000001148.4
ENSMUST00000105411.2 |
Pcbp3
|
poly(rC) binding protein 3 |
| chr7_+_19291070 | 2.64 |
ENSMUST00000108468.3
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chrX_-_111463043 | 2.63 |
ENSMUST00000065976.5
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr8_-_46211284 | 2.60 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr7_-_141443314 | 2.57 |
ENSMUST00000106005.2
|
Lrdd
|
leucine-rich and death domain containing |
| chr17_+_29319183 | 2.57 |
ENSMUST00000114699.1
ENSMUST00000155348.1 |
Pi16
|
peptidase inhibitor 16 |
| chr12_+_111417430 | 2.56 |
ENSMUST00000072646.6
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr12_+_80518990 | 2.55 |
ENSMUST00000021558.6
|
Galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr11_-_120648104 | 2.55 |
ENSMUST00000026134.2
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr13_-_76056996 | 2.53 |
ENSMUST00000056130.4
|
Gpr150
|
G protein-coupled receptor 150 |
| chr2_+_118813995 | 2.52 |
ENSMUST00000134661.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chrX_+_144688907 | 2.52 |
ENSMUST00000112843.1
|
Zcchc16
|
zinc finger, CCHC domain containing 16 |
| chr7_-_45239108 | 2.48 |
ENSMUST00000033063.6
|
Cd37
|
CD37 antigen |
| chr1_-_190979280 | 2.47 |
ENSMUST00000166139.1
|
Vash2
|
vasohibin 2 |
| chr8_-_121907678 | 2.45 |
ENSMUST00000045557.9
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr1_-_43163891 | 2.42 |
ENSMUST00000008280.7
|
Fhl2
|
four and a half LIM domains 2 |
| chr11_-_76509419 | 2.42 |
ENSMUST00000094012.4
|
Abr
|
active BCR-related gene |
| chr14_-_67715585 | 2.40 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr10_-_127195709 | 2.40 |
ENSMUST00000038217.7
ENSMUST00000130855.1 ENSMUST00000116229.1 ENSMUST00000144322.1 |
Dtx3
|
deltex 3 homolog (Drosophila) |
| chr2_+_156840966 | 2.40 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr13_-_55329723 | 2.39 |
ENSMUST00000021941.7
|
Mxd3
|
Max dimerization protein 3 |
| chr6_-_83527986 | 2.37 |
ENSMUST00000121731.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr19_-_41802028 | 2.37 |
ENSMUST00000026150.8
ENSMUST00000177495.1 ENSMUST00000163265.1 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr10_+_60349318 | 2.33 |
ENSMUST00000105459.1
|
4632428N05Rik
|
RIKEN cDNA 4632428N05 gene |
| chr17_+_35059035 | 2.33 |
ENSMUST00000007255.6
ENSMUST00000174493.1 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_+_121760000 | 2.31 |
ENSMUST00000093772.3
|
Zfp651
|
zinc finger protein 651 |
| chr1_+_91366412 | 2.31 |
ENSMUST00000086861.5
|
Fam132b
|
family with sequence similarity 132, member B |
| chr19_-_42431778 | 2.30 |
ENSMUST00000048630.6
|
Crtac1
|
cartilage acidic protein 1 |
| chr1_+_74791516 | 2.29 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr4_-_129121699 | 2.28 |
ENSMUST00000135763.1
ENSMUST00000149763.1 ENSMUST00000164649.1 |
Hpca
|
hippocalcin |
| chr2_+_118814195 | 2.28 |
ENSMUST00000110842.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr14_-_55788810 | 2.27 |
ENSMUST00000022830.6
ENSMUST00000168716.1 ENSMUST00000178399.1 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr15_-_101491509 | 2.26 |
ENSMUST00000023718.7
|
5430421N21Rik
|
RIKEN cDNA 5430421N21 gene |
| chr2_+_69822370 | 2.26 |
ENSMUST00000053087.3
|
Klhl23
|
kelch-like 23 |
| chr14_+_55854115 | 2.25 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr15_+_66670749 | 2.25 |
ENSMUST00000065916.7
|
Tg
|
thyroglobulin |
| chr2_+_181715005 | 2.25 |
ENSMUST00000071585.3
ENSMUST00000148334.1 ENSMUST00000108763.1 |
Oprl1
|
opioid receptor-like 1 |
| chr4_-_151044564 | 2.25 |
ENSMUST00000103204.4
|
Per3
|
period circadian clock 3 |
| chr7_+_78913765 | 2.23 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr4_-_129558355 | 2.23 |
ENSMUST00000167288.1
ENSMUST00000134336.1 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr1_+_75400070 | 2.23 |
ENSMUST00000113589.1
|
Speg
|
SPEG complex locus |
| chr6_+_29433131 | 2.22 |
ENSMUST00000090474.4
|
Flnc
|
filamin C, gamma |
| chr7_+_30421724 | 2.21 |
ENSMUST00000108176.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr8_+_70315759 | 2.21 |
ENSMUST00000165819.2
ENSMUST00000140239.1 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chrX_-_150813637 | 2.21 |
ENSMUST00000112700.1
|
Maged2
|
melanoma antigen, family D, 2 |
| chr2_+_118814237 | 2.21 |
ENSMUST00000028803.7
ENSMUST00000126045.1 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr6_-_125236974 | 2.19 |
ENSMUST00000112281.1
|
Cd27
|
CD27 antigen |
| chr8_+_123332676 | 2.18 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr3_-_67463828 | 2.18 |
ENSMUST00000058981.2
|
Lxn
|
latexin |
| chr7_-_45238794 | 2.18 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chrX_-_111463103 | 2.16 |
ENSMUST00000137712.2
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr9_+_62838767 | 2.14 |
ENSMUST00000034776.6
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr4_-_133498538 | 2.12 |
ENSMUST00000125541.1
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr7_+_43797567 | 2.09 |
ENSMUST00000085461.2
|
Klk8
|
kallikrein related-peptidase 8 |
| chr8_-_25101734 | 2.09 |
ENSMUST00000098866.4
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr17_+_72918298 | 2.08 |
ENSMUST00000024857.6
|
Lbh
|
limb-bud and heart |
| chr3_+_88616133 | 2.07 |
ENSMUST00000176500.1
ENSMUST00000177498.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_91801453 | 2.05 |
ENSMUST00000007949.3
|
Twist2
|
twist basic helix-loop-helix transcription factor 2 |
| chr8_-_87959560 | 2.04 |
ENSMUST00000109655.2
|
Zfp423
|
zinc finger protein 423 |
| chr19_-_4877882 | 2.04 |
ENSMUST00000006626.3
|
Actn3
|
actinin alpha 3 |
| chr10_-_127189981 | 2.03 |
ENSMUST00000019611.7
|
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr4_-_129558387 | 2.03 |
ENSMUST00000067240.4
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr7_+_45434876 | 2.01 |
ENSMUST00000107766.1
|
Gys1
|
glycogen synthase 1, muscle |
| chr13_+_108214389 | 1.99 |
ENSMUST00000022207.8
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr6_-_125236996 | 1.99 |
ENSMUST00000032486.6
|
Cd27
|
CD27 antigen |
| chrX_+_162760427 | 1.98 |
ENSMUST00000112326.1
|
Rbbp7
|
retinoblastoma binding protein 7 |
| chr2_-_92370999 | 1.97 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr2_-_32083783 | 1.96 |
ENSMUST00000056406.6
|
Fam78a
|
family with sequence similarity 78, member A |
| chr5_+_37245792 | 1.96 |
ENSMUST00000031004.7
|
Crmp1
|
collapsin response mediator protein 1 |
| chr1_+_85928483 | 1.96 |
ENSMUST00000027426.4
|
4933407L21Rik
|
RIKEN cDNA 4933407L21 gene |
| chr1_+_85928727 | 1.94 |
ENSMUST00000129392.1
|
4933407L21Rik
|
RIKEN cDNA 4933407L21 gene |
| chrX_+_74329058 | 1.92 |
ENSMUST00000004326.3
|
Plxna3
|
plexin A3 |
| chr8_+_123411424 | 1.92 |
ENSMUST00000071134.3
|
Tubb3
|
tubulin, beta 3 class III |
| chr9_+_22099271 | 1.92 |
ENSMUST00000001384.4
|
Cnn1
|
calponin 1 |
| chr10_+_83722865 | 1.91 |
ENSMUST00000150459.1
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr6_-_83527773 | 1.91 |
ENSMUST00000152029.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chrX_+_135885851 | 1.90 |
ENSMUST00000180025.1
ENSMUST00000068755.7 ENSMUST00000148374.1 |
Bhlhb9
|
basic helix-loop-helix domain containing, class B9 |
| chr2_-_134644019 | 1.90 |
ENSMUST00000110120.1
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr11_+_43528759 | 1.89 |
ENSMUST00000050574.6
|
Ccnjl
|
cyclin J-like |
| chr7_+_4925802 | 1.89 |
ENSMUST00000057612.7
|
Ssc5d
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr12_-_8539545 | 1.88 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr7_-_133123770 | 1.87 |
ENSMUST00000164896.1
ENSMUST00000171968.1 |
Ctbp2
|
C-terminal binding protein 2 |
| chr7_-_133015248 | 1.86 |
ENSMUST00000169570.1
|
Ctbp2
|
C-terminal binding protein 2 |
| chr5_-_67847360 | 1.86 |
ENSMUST00000072971.6
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr17_+_40115358 | 1.86 |
ENSMUST00000061746.7
|
Gm7148
|
predicted gene 7148 |
| chr7_-_24299310 | 1.86 |
ENSMUST00000145131.1
|
Zfp61
|
zinc finger protein 61 |
| chr3_-_90389884 | 1.85 |
ENSMUST00000029541.5
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr9_-_107985863 | 1.85 |
ENSMUST00000048568.4
|
Fam212a
|
family with sequence similarity 212, member A |
| chr17_-_48432723 | 1.85 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr14_+_57524734 | 1.84 |
ENSMUST00000089494.4
|
Il17d
|
interleukin 17D |
| chr6_+_120093348 | 1.83 |
ENSMUST00000112711.2
|
Ninj2
|
ninjurin 2 |
| chr11_+_102604370 | 1.82 |
ENSMUST00000057893.5
|
Fzd2
|
frizzled homolog 2 (Drosophila) |
| chr7_-_133123160 | 1.82 |
ENSMUST00000166439.1
|
Ctbp2
|
C-terminal binding protein 2 |
| chr4_-_129121889 | 1.82 |
ENSMUST00000139450.1
ENSMUST00000125931.1 ENSMUST00000116444.2 |
Hpca
|
hippocalcin |
| chr2_+_155751117 | 1.81 |
ENSMUST00000029140.5
ENSMUST00000132608.1 |
Procr
|
protein C receptor, endothelial |
| chr12_-_101028983 | 1.81 |
ENSMUST00000068411.3
ENSMUST00000085096.3 |
Ccdc88c
|
coiled-coil domain containing 88C |
| chr17_-_24479034 | 1.80 |
ENSMUST00000179163.1
ENSMUST00000070888.6 |
Mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Plag1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 3.2 | 9.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 2.7 | 21.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.3 | 6.8 | GO:0002215 | defense response to nematode(GO:0002215) |
| 2.0 | 7.9 | GO:0051944 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 1.9 | 5.6 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.8 | 5.3 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.7 | 8.6 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 1.7 | 6.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.6 | 9.7 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 1.3 | 1.3 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 1.3 | 6.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.2 | 6.2 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 1.1 | 4.3 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 1.1 | 4.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.1 | 3.2 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 1.0 | 3.9 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.0 | 2.9 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.9 | 3.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.9 | 3.8 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.9 | 3.8 | GO:0035524 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.9 | 7.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.9 | 3.6 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.9 | 4.3 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.9 | 2.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.8 | 8.3 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.8 | 5.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.8 | 2.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.8 | 3.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.8 | 7.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.8 | 1.5 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.8 | 6.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.7 | 2.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.7 | 2.2 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.7 | 2.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.7 | 2.2 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.7 | 8.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.7 | 5.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.7 | 1.4 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.7 | 8.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.7 | 1.3 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 0.6 | 3.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.6 | 4.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.6 | 7.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.6 | 2.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 1.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.6 | 2.3 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.6 | 3.9 | GO:0030421 | defecation(GO:0030421) |
| 0.6 | 8.4 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.6 | 1.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.6 | 6.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.5 | 4.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.5 | 2.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.5 | 4.6 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.5 | 5.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.5 | 2.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 2.0 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.5 | 2.0 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 1.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 1.4 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.5 | 1.9 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.5 | 3.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.5 | 1.4 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.5 | 1.8 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.5 | 3.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 0.9 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.4 | 2.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 1.8 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 5.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.4 | 1.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 | 1.3 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.4 | 1.7 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.6 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.4 | 6.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.4 | 3.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 0.8 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.4 | 3.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 1.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 1.9 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 3.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 1.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 1.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.4 | 4.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.4 | 1.8 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.4 | 4.3 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.4 | 6.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 5.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 9.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.3 | 2.7 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.3 | 1.7 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) positive regulation of neurotrophin production(GO:0032901) |
| 0.3 | 4.3 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) negative regulation of calcium ion import(GO:0090281) |
| 0.3 | 1.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.3 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.3 | 1.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.6 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.3 | 1.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.3 | 5.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.3 | 1.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 0.9 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.3 | 2.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 0.9 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.3 | 4.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 1.1 | GO:1902202 | proteoglycan catabolic process(GO:0030167) regulation of endothelial tube morphogenesis(GO:1901509) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 3.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 0.8 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.3 | 2.2 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.3 | 2.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 6.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 0.5 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.3 | 2.9 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.3 | 1.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.3 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 0.8 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 1.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.3 | 1.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 0.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 1.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 1.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 1.7 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.2 | 0.7 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 0.5 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 3.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 4.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 2.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 0.5 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.2 | 2.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 0.7 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.2 | 2.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.9 | GO:0010958 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 4.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.3 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.2 | 0.6 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 2.7 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 12.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 1.2 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.2 | 2.9 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.2 | 1.3 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.2 | 1.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 0.2 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.2 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 2.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.6 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 1.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 4.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 2.1 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.2 | 0.7 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 1.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 0.9 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.2 | 4.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.9 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 0.5 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.2 | 2.0 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.5 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.7 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 1.0 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 1.5 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.2 | 0.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 0.6 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 1.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 7.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 2.4 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 1.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.4 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 1.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.7 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.4 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.1 | 0.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 1.2 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 1.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 2.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.3 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 4.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 5.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.7 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.4 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 1.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.9 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.7 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.5 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.8 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 1.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.3 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.8 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 1.1 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.1 | 0.3 | GO:0002865 | type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 0.1 | 0.7 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.9 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.4 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.5 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 1.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 1.5 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 1.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 1.5 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.1 | 0.2 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 1.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 2.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0048686 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 0.9 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.5 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 2.4 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 1.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 2.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 1.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 1.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 0.5 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.2 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.1 | 0.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 1.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.2 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.4 | GO:1903365 | positive regulation of synapse structural plasticity(GO:0051835) regulation of fear response(GO:1903365) regulation of AMPA glutamate receptor clustering(GO:1904717) regulation of behavioral fear response(GO:2000822) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 1.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.8 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.1 | 0.3 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.5 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 0.4 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.1 | 1.4 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 6.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.8 | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.1 | 5.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 1.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.2 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 2.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.2 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 2.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 1.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 1.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.9 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.4 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.1 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.2 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 0.9 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 1.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.4 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 0.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.4 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 3.0 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 3.1 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 3.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 3.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 2.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 1.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 5.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 0.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.6 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.0 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 1.2 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 2.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 1.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 1.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.4 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0000056 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.3 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.4 | GO:0071554 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.6 | GO:0032292 | Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0046600 | histone H2A-K119 monoubiquitination(GO:0036353) negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.7 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 1.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:1902416 | regulation of mRNA binding(GO:1902415) positive regulation of mRNA binding(GO:1902416) regulation of RNA binding(GO:1905214) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.5 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.4 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) |
| 0.0 | 0.6 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.0 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.6 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.6 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.7 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.1 | GO:0045606 | regulation of water loss via skin(GO:0033561) positive regulation of epidermal cell differentiation(GO:0045606) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 2.2 | 6.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.4 | 1.4 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 1.1 | 5.6 | GO:0031523 | Myb complex(GO:0031523) |
| 1.1 | 4.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.0 | 3.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.7 | 9.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.7 | 2.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.7 | 3.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.7 | 10.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.6 | 9.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 8.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.5 | 3.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 8.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 2.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 1.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 2.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.4 | 2.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 8.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.4 | 9.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 1.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 0.7 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.3 | 5.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 2.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 1.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 2.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.3 | 0.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 5.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 8.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 0.9 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 1.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 0.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 1.3 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.2 | 3.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 5.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 2.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 1.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 4.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 1.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 3.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.0 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 10.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 16.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 0.9 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 0.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.1 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 6.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 3.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 2.0 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 2.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.0 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 2.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.3 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.1 | 1.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 5.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.3 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.3 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 13.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 3.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.4 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 5.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 4.0 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.9 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 2.2 | 8.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.6 | 6.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.3 | 3.8 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 1.2 | 3.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.0 | 9.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.9 | 3.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.9 | 4.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.9 | 4.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.8 | 8.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.7 | 7.9 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.6 | 5.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.6 | 2.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.6 | 21.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.6 | 6.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.6 | 1.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.5 | 1.6 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.5 | 4.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.5 | 1.6 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.5 | 5.6 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.5 | 5.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 3.7 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.4 | 1.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.4 | 6.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.4 | 3.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 1.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 2.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 3.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 4.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 1.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.3 | 5.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 16.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 1.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.3 | 5.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 1.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 3.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 0.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 10.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.3 | 1.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 1.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 1.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 3.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 4.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 2.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.3 | 2.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 1.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.3 | 2.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 0.8 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.3 | 1.8 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.3 | 3.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 2.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 7.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 1.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 6.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 1.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.6 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.2 | 0.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 6.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 2.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.8 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 1.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 3.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 6.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 1.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 6.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 4.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 3.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.0 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 9.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 6.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 3.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.4 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.1 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 6.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 3.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 2.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single guanine insertion binding(GO:0032142) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 3.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 5.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 6.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 2.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 3.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 5.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 3.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 2.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.9 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 3.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 6.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 2.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 14.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.3 | 3.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 8.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 11.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 10.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 10.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 11.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 1.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 9.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 14.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 4.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.6 | 9.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 15.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.6 | 10.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 4.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 5.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 15.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 15.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.3 | 2.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 3.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 0.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 7.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 1.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 5.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 5.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 5.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 3.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 3.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 11.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 5.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 2.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 3.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 6.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 5.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 4.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 5.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 3.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 4.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |