Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Prrx1_Isx_Prrxl1
Z-value: 0.96
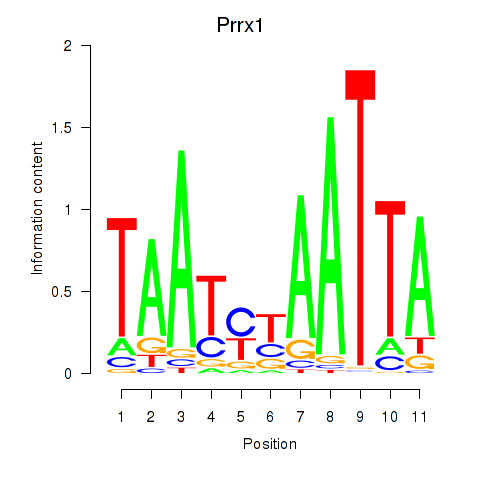
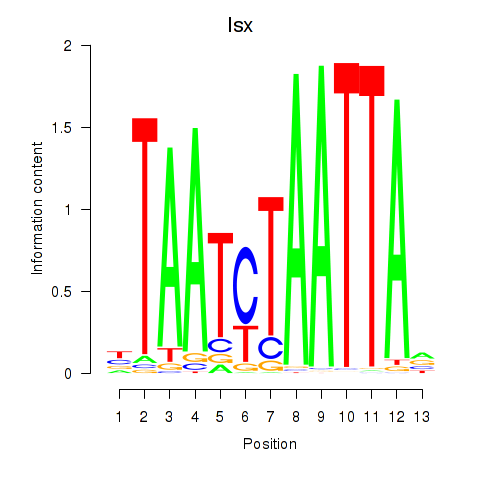
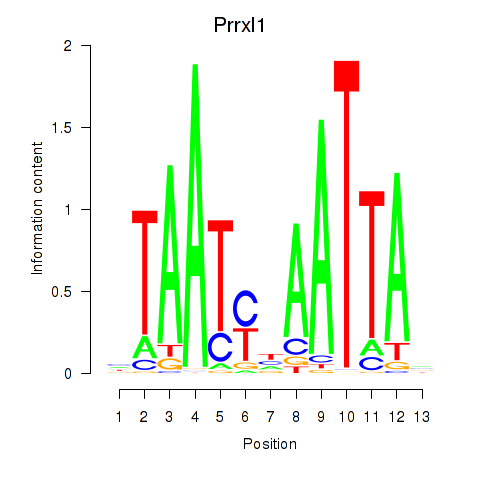
Transcription factors associated with Prrx1_Isx_Prrxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prrx1
|
ENSMUSG00000026586.10 | paired related homeobox 1 |
|
Isx
|
ENSMUSG00000031621.3 | intestine specific homeobox |
|
Prrxl1
|
ENSMUSG00000041730.7 | paired related homeobox protein-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isx | mm10_v2_chr8_+_74873540_74873615 | -0.20 | 2.4e-01 | Click! |
| Prrx1 | mm10_v2_chr1_-_163289214_163289235 | -0.12 | 5.0e-01 | Click! |
Activity profile of Prrx1_Isx_Prrxl1 motif
Sorted Z-values of Prrx1_Isx_Prrxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_99322943 | 1.65 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr8_+_21565368 | 1.55 |
ENSMUST00000084040.2
|
Gm14850
|
predicted gene 14850 |
| chr16_-_45693658 | 1.38 |
ENSMUST00000114562.2
ENSMUST00000036617.7 |
Tmprss7
|
transmembrane serine protease 7 |
| chr8_+_21025545 | 1.27 |
ENSMUST00000076754.2
|
Defa21
|
defensin, alpha, 21 |
| chr4_-_87806276 | 1.24 |
ENSMUST00000148059.1
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr1_-_162898665 | 1.21 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr4_-_87806296 | 1.17 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr6_+_37870786 | 1.15 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chrM_+_11734 | 1.13 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr8_+_21391811 | 1.04 |
ENSMUST00000120874.3
|
Gm21002
|
predicted gene, 21002 |
| chr8_+_21529031 | 0.97 |
ENSMUST00000084041.3
|
Gm15308
|
predicted gene 15308 |
| chr8_+_21297439 | 0.95 |
ENSMUST00000098892.4
|
Defa5
|
defensin, alpha, 5 |
| chr8_+_21427456 | 0.91 |
ENSMUST00000095424.4
|
Gm21498
|
predicted gene, 21498 |
| chr2_-_67194695 | 0.90 |
ENSMUST00000147939.1
|
Gm13598
|
predicted gene 13598 |
| chr5_-_87091150 | 0.89 |
ENSMUST00000154455.1
|
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr3_+_146121655 | 0.88 |
ENSMUST00000039450.4
|
Mcoln3
|
mucolipin 3 |
| chr12_+_79297345 | 0.86 |
ENSMUST00000079533.5
ENSMUST00000171210.1 |
Rad51b
|
RAD51 homolog B |
| chr1_+_87803638 | 0.81 |
ENSMUST00000077772.5
ENSMUST00000177757.1 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chr8_-_105568298 | 0.81 |
ENSMUST00000005849.5
|
Agrp
|
agouti related protein |
| chr13_+_104229366 | 0.80 |
ENSMUST00000022227.6
|
Cenpk
|
centromere protein K |
| chr8_+_21509258 | 0.79 |
ENSMUST00000084042.3
|
Defa20
|
defensin, alpha, 20 |
| chr8_-_3854309 | 0.75 |
ENSMUST00000033888.4
|
Cd209e
|
CD209e antigen |
| chr8_+_21665797 | 0.73 |
ENSMUST00000075268.4
|
Gm15315
|
predicted gene 15315 |
| chr11_+_94936224 | 0.73 |
ENSMUST00000001547.7
|
Col1a1
|
collagen, type I, alpha 1 |
| chr2_-_160619971 | 0.73 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr2_-_164638789 | 0.72 |
ENSMUST00000109336.1
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chr9_-_71896047 | 0.69 |
ENSMUST00000184448.1
|
Tcf12
|
transcription factor 12 |
| chr4_+_103143052 | 0.64 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr19_+_39287074 | 0.63 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr8_-_21703642 | 0.60 |
ENSMUST00000071886.6
|
AY761184
|
cDNA sequence AY761184 |
| chr11_-_12412136 | 0.59 |
ENSMUST00000174874.1
|
Cobl
|
cordon-bleu WH2 repeat |
| chr7_-_68353182 | 0.57 |
ENSMUST00000123509.1
ENSMUST00000129965.1 |
Gm16158
|
predicted gene 16158 |
| chr14_-_104522615 | 0.56 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr9_-_42399709 | 0.54 |
ENSMUST00000160940.1
|
Tecta
|
tectorin alpha |
| chr14_+_53704007 | 0.53 |
ENSMUST00000103664.4
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr9_-_42399915 | 0.52 |
ENSMUST00000042190.7
|
Tecta
|
tectorin alpha |
| chr4_+_5724304 | 0.52 |
ENSMUST00000108380.1
|
Fam110b
|
family with sequence similarity 110, member B |
| chrX_-_162964557 | 0.52 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr12_+_117843489 | 0.51 |
ENSMUST00000021592.9
|
Cdca7l
|
cell division cycle associated 7 like |
| chr3_-_59220150 | 0.51 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr2_+_14873656 | 0.49 |
ENSMUST00000114718.1
ENSMUST00000114719.1 |
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr18_+_37320374 | 0.47 |
ENSMUST00000078271.2
|
Pcdhb5
|
protocadherin beta 5 |
| chr1_-_152625212 | 0.46 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr5_+_109940426 | 0.46 |
ENSMUST00000170826.1
|
Gm15446
|
predicted gene 15446 |
| chr9_-_59353430 | 0.45 |
ENSMUST00000026265.6
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr7_+_100159241 | 0.45 |
ENSMUST00000032967.3
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr9_-_39604124 | 0.45 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr10_-_6980376 | 0.44 |
ENSMUST00000105617.1
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_-_162898484 | 0.44 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr14_+_53649387 | 0.43 |
ENSMUST00000103660.2
|
Trav15-2-dv6-2
|
T cell receptor alpha variable 15-2-DV6-2 |
| chr2_-_26671282 | 0.43 |
ENSMUST00000028283.1
|
Lcn4
|
lipocalin 4 |
| chr4_+_35152056 | 0.41 |
ENSMUST00000058595.6
|
Ifnk
|
interferon kappa |
| chr3_+_3634145 | 0.39 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr7_+_29170345 | 0.38 |
ENSMUST00000033886.7
|
Ggn
|
gametogenetin |
| chr7_-_6730412 | 0.38 |
ENSMUST00000051209.4
|
Peg3
|
paternally expressed 3 |
| chr8_-_21594508 | 0.37 |
ENSMUST00000110749.3
|
Gm7861
|
predicted gene 7861 |
| chr9_+_118478182 | 0.37 |
ENSMUST00000111763.1
|
Eomes
|
eomesodermin homolog (Xenopus laevis) |
| chr15_+_81744848 | 0.37 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr8_-_122360746 | 0.37 |
ENSMUST00000044123.1
|
Trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr2_-_72986716 | 0.36 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr13_+_67833235 | 0.36 |
ENSMUST00000060609.7
|
Gm10037
|
predicted gene 10037 |
| chr17_-_32822200 | 0.36 |
ENSMUST00000179695.1
|
Zfp799
|
zinc finger protein 799 |
| chr4_+_74242468 | 0.36 |
ENSMUST00000077851.3
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr8_+_94172618 | 0.35 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr13_+_49504774 | 0.35 |
ENSMUST00000051504.7
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr11_-_46707059 | 0.35 |
ENSMUST00000055102.6
ENSMUST00000125008.1 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chrX_-_94212638 | 0.35 |
ENSMUST00000113922.1
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr3_+_32436376 | 0.34 |
ENSMUST00000108242.1
|
Pik3ca
|
phosphatidylinositol 3-kinase, catalytic, alpha polypeptide |
| chr3_+_66219909 | 0.34 |
ENSMUST00000029421.5
|
Ptx3
|
pentraxin related gene |
| chr9_+_118478344 | 0.34 |
ENSMUST00000035020.8
|
Eomes
|
eomesodermin homolog (Xenopus laevis) |
| chr17_-_37483543 | 0.34 |
ENSMUST00000016427.4
ENSMUST00000171139.2 |
H2-M2
|
histocompatibility 2, M region locus 2 |
| chr15_+_85510812 | 0.34 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr4_+_102430047 | 0.33 |
ENSMUST00000172616.1
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_+_155751117 | 0.33 |
ENSMUST00000029140.5
ENSMUST00000132608.1 |
Procr
|
protein C receptor, endothelial |
| chr7_+_45621805 | 0.33 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr18_+_4375583 | 0.33 |
ENSMUST00000025077.6
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr10_-_76110956 | 0.33 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr1_-_24612700 | 0.33 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr3_+_152165374 | 0.32 |
ENSMUST00000181854.1
|
D630002J18Rik
|
RIKEN cDNA D630002J18 gene |
| chr7_+_126862431 | 0.32 |
ENSMUST00000132808.1
|
Hirip3
|
HIRA interacting protein 3 |
| chr1_-_150466165 | 0.32 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr2_-_73452666 | 0.31 |
ENSMUST00000151939.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_170308802 | 0.30 |
ENSMUST00000056991.5
|
1700015E13Rik
|
RIKEN cDNA 1700015E13 gene |
| chr13_+_102693522 | 0.29 |
ENSMUST00000022124.3
ENSMUST00000171267.1 ENSMUST00000167144.1 ENSMUST00000170878.1 |
Cd180
|
CD180 antigen |
| chr11_-_30268169 | 0.29 |
ENSMUST00000006629.7
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr3_+_41742615 | 0.29 |
ENSMUST00000146165.1
ENSMUST00000119572.1 ENSMUST00000108065.2 ENSMUST00000120167.1 ENSMUST00000026867.7 ENSMUST00000026868.7 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr6_+_97991776 | 0.29 |
ENSMUST00000043628.6
|
Mitf
|
microphthalmia-associated transcription factor |
| chr3_-_41742471 | 0.28 |
ENSMUST00000026866.8
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr5_+_15516489 | 0.28 |
ENSMUST00000178227.1
|
Gm21847
|
predicted gene, 21847 |
| chr4_+_102589687 | 0.28 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr1_+_58210397 | 0.28 |
ENSMUST00000040442.5
|
Aox4
|
aldehyde oxidase 4 |
| chr2_-_45110336 | 0.28 |
ENSMUST00000028229.6
ENSMUST00000152232.1 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_122915987 | 0.27 |
ENSMUST00000098333.4
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr15_-_9140374 | 0.27 |
ENSMUST00000096482.3
ENSMUST00000110585.2 |
Skp2
|
S-phase kinase-associated protein 2 (p45) |
| chr19_-_32196393 | 0.27 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr18_-_38866702 | 0.27 |
ENSMUST00000115582.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr4_+_34893772 | 0.27 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr18_+_37411674 | 0.26 |
ENSMUST00000051126.2
|
Pcdhb10
|
protocadherin beta 10 |
| chr4_+_102741287 | 0.26 |
ENSMUST00000097948.2
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr9_+_92457369 | 0.26 |
ENSMUST00000034941.7
|
Plscr4
|
phospholipid scramblase 4 |
| chr16_+_92478743 | 0.26 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chr3_-_49757257 | 0.26 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr5_-_124095749 | 0.26 |
ENSMUST00000031354.4
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr1_+_107422681 | 0.26 |
ENSMUST00000112710.1
ENSMUST00000086690.4 |
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr18_-_39487096 | 0.26 |
ENSMUST00000097592.2
ENSMUST00000115571.1 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr3_+_122895072 | 0.26 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr6_-_37442095 | 0.26 |
ENSMUST00000041093.5
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr7_+_26757153 | 0.25 |
ENSMUST00000077855.6
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr1_+_180111339 | 0.25 |
ENSMUST00000145181.1
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_125136692 | 0.25 |
ENSMUST00000099452.2
|
Ctxn2
|
cortexin 2 |
| chr11_-_94782703 | 0.25 |
ENSMUST00000100554.1
|
Tmem92
|
transmembrane protein 92 |
| chrX_+_99975570 | 0.25 |
ENSMUST00000113779.1
ENSMUST00000113776.1 ENSMUST00000113775.1 ENSMUST00000113780.1 ENSMUST00000113778.1 ENSMUST00000113781.1 ENSMUST00000113783.1 ENSMUST00000071453.2 ENSMUST00000113777.1 |
Eda
|
ectodysplasin-A |
| chr3_+_66985947 | 0.25 |
ENSMUST00000161726.1
ENSMUST00000160504.1 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_+_95557494 | 0.25 |
ENSMUST00000079223.4
ENSMUST00000177394.1 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr1_+_10056922 | 0.24 |
ENSMUST00000149214.1
|
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr17_+_21690766 | 0.24 |
ENSMUST00000097384.1
|
Gm10509
|
predicted gene 10509 |
| chr13_+_96388294 | 0.23 |
ENSMUST00000099295.4
|
Poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr11_-_99374895 | 0.23 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr18_-_47333311 | 0.23 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_+_41966058 | 0.23 |
ENSMUST00000108026.2
|
Gm20938
|
predicted gene, 20938 |
| chr13_+_19342154 | 0.23 |
ENSMUST00000103566.3
|
Tcrg-C4
|
T cell receptor gamma, constant 4 |
| chr14_-_118923070 | 0.23 |
ENSMUST00000047208.5
|
Dzip1
|
DAZ interacting protein 1 |
| chr4_-_41517326 | 0.23 |
ENSMUST00000030152.6
ENSMUST00000095126.4 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr14_+_84443553 | 0.23 |
ENSMUST00000071370.5
|
Pcdh17
|
protocadherin 17 |
| chr1_-_150993051 | 0.23 |
ENSMUST00000074783.5
ENSMUST00000137197.2 |
Hmcn1
|
hemicentin 1 |
| chr7_-_5413145 | 0.23 |
ENSMUST00000108569.2
|
Vmn1r58
|
vomeronasal 1 receptor 58 |
| chr4_+_126556935 | 0.23 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr16_-_90810365 | 0.22 |
ENSMUST00000140920.1
|
Urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr11_-_94782500 | 0.22 |
ENSMUST00000162809.2
|
Tmem92
|
transmembrane protein 92 |
| chr11_-_94297067 | 0.22 |
ENSMUST00000132623.2
|
Luc7l3
|
LUC7-like 3 (S. cerevisiae) |
| chr14_-_51195923 | 0.22 |
ENSMUST00000051274.1
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chrX_+_10252361 | 0.22 |
ENSMUST00000115528.2
|
Otc
|
ornithine transcarbamylase |
| chr11_-_64436653 | 0.22 |
ENSMUST00000177999.1
|
F930015N05Rik
|
RIKEN cDNA F930015N05 gene |
| chr3_+_19989151 | 0.21 |
ENSMUST00000173779.1
|
Cp
|
ceruloplasmin |
| chr10_+_69925484 | 0.21 |
ENSMUST00000182692.1
ENSMUST00000092433.5 |
Ank3
|
ankyrin 3, epithelial |
| chr9_+_65361049 | 0.21 |
ENSMUST00000147185.1
|
Gm514
|
predicted gene 514 |
| chr11_-_54249640 | 0.21 |
ENSMUST00000019060.5
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chrX_+_10252305 | 0.21 |
ENSMUST00000049910.6
|
Otc
|
ornithine transcarbamylase |
| chr13_-_27513206 | 0.21 |
ENSMUST00000018389.3
ENSMUST00000110350.1 |
Prl8a8
|
prolactin family 8, subfamily a, member 81 |
| chr15_-_100424092 | 0.21 |
ENSMUST00000154676.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chrX_+_56963325 | 0.21 |
ENSMUST00000096431.3
|
Gpr112
|
G protein-coupled receptor 112 |
| chr8_+_21065038 | 0.21 |
ENSMUST00000078121.3
|
Gm10104
|
predicted gene 10104 |
| chr14_-_67072465 | 0.21 |
ENSMUST00000089230.5
|
Ppp2r2a
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform |
| chr10_+_69925766 | 0.20 |
ENSMUST00000182269.1
ENSMUST00000183261.1 ENSMUST00000183074.1 |
Ank3
|
ankyrin 3, epithelial |
| chr16_+_16870829 | 0.20 |
ENSMUST00000131063.1
|
Top3b
|
topoisomerase (DNA) III beta |
| chr11_+_104231573 | 0.20 |
ENSMUST00000132977.1
ENSMUST00000132245.1 ENSMUST00000100347.4 |
Mapt
|
microtubule-associated protein tau |
| chr10_-_13324160 | 0.20 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr7_-_101581161 | 0.20 |
ENSMUST00000063920.2
|
Art2b
|
ADP-ribosyltransferase 2b |
| chr10_+_79704499 | 0.20 |
ENSMUST00000067036.5
ENSMUST00000178383.1 |
Bsg
|
basigin |
| chr15_-_37459327 | 0.20 |
ENSMUST00000119730.1
ENSMUST00000120746.1 |
Ncald
|
neurocalcin delta |
| chrM_+_9870 | 0.20 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr16_+_33684460 | 0.20 |
ENSMUST00000152782.1
ENSMUST00000179453.1 |
Heg1
|
HEG homolog 1 (zebrafish) |
| chr11_+_73350839 | 0.20 |
ENSMUST00000120137.1
|
Olfr20
|
olfactory receptor 20 |
| chr16_-_22811399 | 0.20 |
ENSMUST00000040592.4
|
Crygs
|
crystallin, gamma S |
| chr7_+_43351378 | 0.20 |
ENSMUST00000012798.7
ENSMUST00000122423.1 ENSMUST00000121494.1 |
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr2_-_17460610 | 0.19 |
ENSMUST00000145492.1
|
Nebl
|
nebulette |
| chr14_+_52824340 | 0.19 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr11_+_104231465 | 0.19 |
ENSMUST00000145227.1
|
Mapt
|
microtubule-associated protein tau |
| chr10_+_69925954 | 0.19 |
ENSMUST00000181974.1
ENSMUST00000182795.1 ENSMUST00000182437.1 |
Ank3
|
ankyrin 3, epithelial |
| chr13_+_23034294 | 0.19 |
ENSMUST00000074252.2
|
Vmn1r214
|
vomeronasal 1 receptor 214 |
| chr16_-_16829276 | 0.19 |
ENSMUST00000023468.5
|
Spag6
|
sperm associated antigen 6 |
| chr12_+_109546409 | 0.19 |
ENSMUST00000143847.1
|
Meg3
|
maternally expressed 3 |
| chr11_-_99543158 | 0.19 |
ENSMUST00000107443.1
ENSMUST00000074253.3 |
Krt40
|
keratin 40 |
| chr6_+_36388055 | 0.18 |
ENSMUST00000172278.1
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr1_-_130661613 | 0.18 |
ENSMUST00000027657.7
|
C4bp
|
complement component 4 binding protein |
| chr4_+_62525369 | 0.18 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chrX_+_163911401 | 0.18 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_+_167598450 | 0.18 |
ENSMUST00000111386.1
ENSMUST00000111384.1 |
Rxrg
|
retinoid X receptor gamma |
| chr17_-_45686899 | 0.18 |
ENSMUST00000156254.1
|
Tmem63b
|
transmembrane protein 63b |
| chrX_-_134111852 | 0.18 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr2_+_68104671 | 0.18 |
ENSMUST00000042456.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr4_+_126556994 | 0.18 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr14_-_110755100 | 0.18 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr12_-_54999102 | 0.18 |
ENSMUST00000173529.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr7_-_14446570 | 0.18 |
ENSMUST00000063509.4
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr4_-_58499398 | 0.17 |
ENSMUST00000107570.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chrX_+_139610612 | 0.17 |
ENSMUST00000113026.1
|
Rnf128
|
ring finger protein 128 |
| chr7_+_29170204 | 0.17 |
ENSMUST00000098609.2
|
Ggn
|
gametogenetin |
| chr11_-_12027958 | 0.17 |
ENSMUST00000109654.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr13_+_93304799 | 0.17 |
ENSMUST00000080127.5
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr6_+_30723541 | 0.17 |
ENSMUST00000115127.1
|
Mest
|
mesoderm specific transcript |
| chr16_+_45224315 | 0.17 |
ENSMUST00000102802.3
ENSMUST00000063654.4 |
Btla
|
B and T lymphocyte associated |
| chr2_+_119047116 | 0.17 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chr2_-_89024099 | 0.17 |
ENSMUST00000099799.2
|
Olfr1217
|
olfactory receptor 1217 |
| chr9_+_105642957 | 0.17 |
ENSMUST00000065778.6
|
Pik3r4
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 4, p150 |
| chr9_+_72806874 | 0.17 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr2_+_10047838 | 0.16 |
ENSMUST00000181588.1
|
C630004M23Rik
|
RIKEN cDNA C630004M23 gene |
| chr7_+_102441685 | 0.16 |
ENSMUST00000033283.9
|
Rrm1
|
ribonucleotide reductase M1 |
| chr13_-_4609122 | 0.16 |
ENSMUST00000110691.3
ENSMUST00000091848.5 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr1_+_180101144 | 0.16 |
ENSMUST00000133890.1
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_-_87535113 | 0.16 |
ENSMUST00000120150.1
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr18_+_69593361 | 0.16 |
ENSMUST00000114978.2
ENSMUST00000114977.1 |
Tcf4
|
transcription factor 4 |
| chr4_-_134767940 | 0.16 |
ENSMUST00000037828.6
|
Ldlrap1
|
low density lipoprotein receptor adaptor protein 1 |
| chr12_-_72408934 | 0.16 |
ENSMUST00000078505.7
|
Rtn1
|
reticulon 1 |
| chr18_+_37421418 | 0.16 |
ENSMUST00000053073.4
|
Pcdhb11
|
protocadherin beta 11 |
| chr3_+_32515295 | 0.15 |
ENSMUST00000029203.7
|
Zfp639
|
zinc finger protein 639 |
| chr7_-_79848191 | 0.15 |
ENSMUST00000107392.1
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr5_+_95106938 | 0.15 |
ENSMUST00000163216.1
|
Gm7942
|
predicted gene 7942 |
| chr5_-_151190154 | 0.15 |
ENSMUST00000062015.8
ENSMUST00000110483.2 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr1_-_9299238 | 0.15 |
ENSMUST00000140295.1
|
Sntg1
|
syntrophin, gamma 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prrx1_Isx_Prrxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 0.7 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 | 1.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 2.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.5 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.4 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.3 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 1.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:0060296 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.1 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.1 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.8 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 1.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.2 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0014878 | regulation of muscle atrophy(GO:0014735) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.4 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.1 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 1.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:2000471 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.7 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.0 | GO:0016259 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.2 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 0.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.6 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.8 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0016623 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 1.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |