Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rarb
Z-value: 1.20
Transcription factors associated with Rarb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarb
|
ENSMUSG00000017491.8 | retinoic acid receptor, beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarb | mm10_v2_chr14_-_16575456_16575501 | -0.04 | 8.1e-01 | Click! |
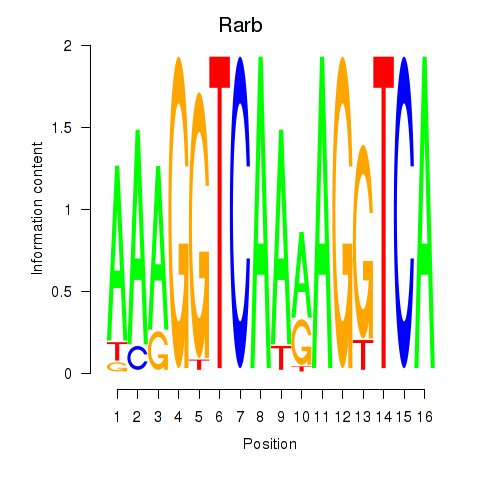
Activity profile of Rarb motif
Sorted Z-values of Rarb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_85820965 | 15.85 |
ENSMUST00000032074.3
|
Cml5
|
camello-like 5 |
| chr5_-_87254804 | 8.07 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr6_-_85820936 | 6.16 |
ENSMUST00000174143.1
|
Gm11128
|
predicted gene 11128 |
| chr6_-_138073196 | 5.20 |
ENSMUST00000050132.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr9_-_46235631 | 4.56 |
ENSMUST00000118649.1
|
Apoc3
|
apolipoprotein C-III |
| chr5_-_87092546 | 4.32 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr17_+_12584183 | 3.89 |
ENSMUST00000046959.7
|
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr8_+_36489191 | 3.43 |
ENSMUST00000171777.1
|
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr5_-_87140318 | 2.92 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_+_76904475 | 2.32 |
ENSMUST00000142166.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr5_+_87000838 | 2.24 |
ENSMUST00000031186.7
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr19_+_28990476 | 2.13 |
ENSMUST00000050148.3
|
Cdc37l1
|
cell division cycle 37-like 1 |
| chr1_+_4807823 | 1.79 |
ENSMUST00000027036.4
ENSMUST00000119612.2 ENSMUST00000137887.1 ENSMUST00000115529.1 ENSMUST00000150971.1 |
Lypla1
|
lysophospholipase 1 |
| chr5_-_86906937 | 1.64 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr10_-_126901315 | 1.38 |
ENSMUST00000026504.5
ENSMUST00000168520.1 |
Xrcc6bp1
|
XRCC6 binding protein 1 |
| chr11_+_76904513 | 1.17 |
ENSMUST00000072633.3
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr19_+_44992127 | 1.05 |
ENSMUST00000179305.1
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr12_+_7977640 | 0.79 |
ENSMUST00000171271.1
ENSMUST00000037811.6 ENSMUST00000037520.7 |
Apob
|
apolipoprotein B |
| chr3_+_154711125 | 0.70 |
ENSMUST00000051862.6
|
4922501L14Rik
|
RIKEN cDNA 4922501L14 gene |
| chr10_+_43524080 | 0.64 |
ENSMUST00000057649.6
|
Gm9803
|
predicted gene 9803 |
| chr3_+_108186332 | 0.58 |
ENSMUST00000050909.6
ENSMUST00000106659.2 ENSMUST00000106656.1 ENSMUST00000106661.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr9_+_56937462 | 0.55 |
ENSMUST00000034827.8
|
Imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr2_+_3770673 | 0.47 |
ENSMUST00000177037.1
|
Fam107b
|
family with sequence similarity 107, member B |
| chr11_+_83409137 | 0.44 |
ENSMUST00000021022.3
|
Rasl10b
|
RAS-like, family 10, member B |
| chr11_+_83409655 | 0.35 |
ENSMUST00000175848.1
ENSMUST00000108140.3 |
Rasl10b
|
RAS-like, family 10, member B |
| chr5_+_92897981 | 0.30 |
ENSMUST00000113051.2
|
Shroom3
|
shroom family member 3 |
| chr4_+_128058962 | 0.30 |
ENSMUST00000184063.1
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr10_-_92722356 | 0.25 |
ENSMUST00000020163.6
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated gene 1 |
| chr17_+_37045963 | 0.22 |
ENSMUST00000025338.9
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr17_+_37045980 | 0.20 |
ENSMUST00000174456.1
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr11_+_105178765 | 0.19 |
ENSMUST00000106939.2
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr4_+_156331103 | 0.09 |
ENSMUST00000074107.6
ENSMUST00000096792.5 |
Vmn2r-ps159
|
vomeronasal 2, receptor, pseudogene 159 |
| chr8_+_106683052 | 0.03 |
ENSMUST00000048359.4
|
Tango6
|
transport and golgi organization 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.3 | 3.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.8 | GO:0002084 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 3.9 | GO:0051608 | histamine transport(GO:0051608) |
| 0.2 | 0.8 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 5.2 | GO:0015833 | peptide transport(GO:0015833) |
| 0.0 | 2.1 | GO:0006457 | protein folding(GO:0006457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 11.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 1.1 | 4.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.6 | 3.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 19.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.6 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 1.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 5.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 4.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 1.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |