Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
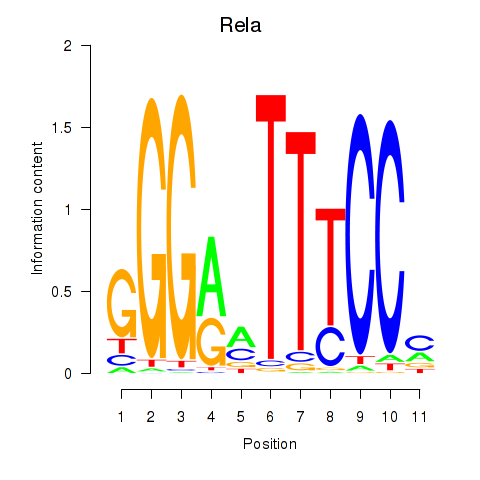
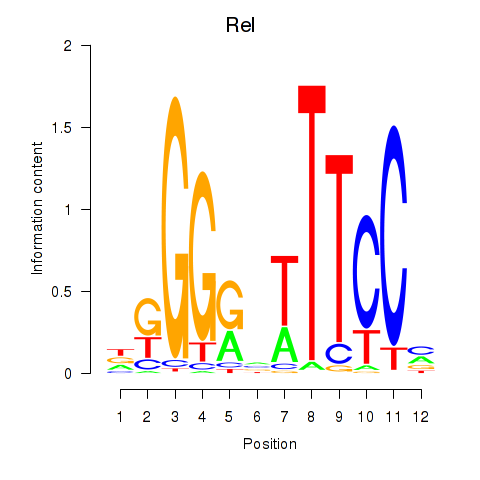
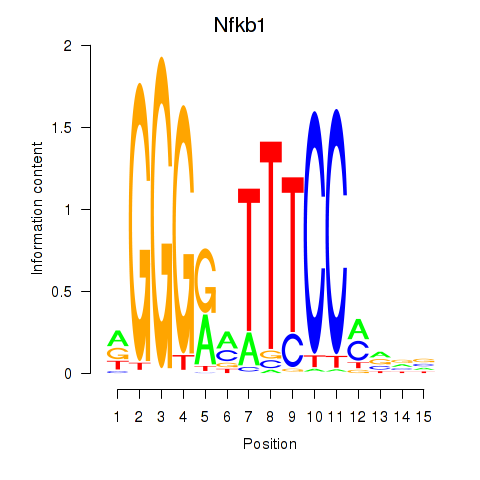
Results for Rela_Rel_Nfkb1
Z-value: 0.96
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSMUSG00000024927.7 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
|
Rel
|
ENSMUSG00000020275.8 | reticuloendotheliosis oncogene |
|
Nfkb1
|
ENSMUSG00000028163.11 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rel | mm10_v2_chr11_-_23770953_23771028 | 0.47 | 3.9e-03 | Click! |
| Nfkb1 | mm10_v2_chr3_-_135691512_135691564 | 0.41 | 1.3e-02 | Click! |
| Rela | mm10_v2_chr19_+_5637475_5637486 | 0.21 | 2.1e-01 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90891234 | 4.37 |
ENSMUST00000031327.8
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr17_+_37193889 | 3.33 |
ENSMUST00000038844.6
|
Ubd
|
ubiquitin D |
| chr1_+_74791516 | 2.88 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr11_-_82179808 | 1.94 |
ENSMUST00000108189.2
ENSMUST00000021043.4 |
Ccl1
|
chemokine (C-C motif) ligand 1 |
| chr15_-_78495059 | 1.74 |
ENSMUST00000089398.1
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr12_+_111166485 | 1.64 |
ENSMUST00000139162.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr11_-_83530505 | 1.57 |
ENSMUST00000035938.2
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr14_-_45529964 | 1.52 |
ENSMUST00000150660.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr7_+_26061495 | 1.42 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr5_-_92348871 | 1.39 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_-_172590463 | 1.33 |
ENSMUST00000065679.6
|
Slamf8
|
SLAM family member 8 |
| chr18_+_60803838 | 1.26 |
ENSMUST00000050487.8
ENSMUST00000097563.2 ENSMUST00000167610.1 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr9_-_14615473 | 1.22 |
ENSMUST00000162901.1
|
Amotl1
|
angiomotin-like 1 |
| chr4_+_150927918 | 1.21 |
ENSMUST00000139826.1
ENSMUST00000116257.1 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr5_+_7960445 | 1.19 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chr17_+_24878724 | 1.16 |
ENSMUST00000050714.6
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr8_-_61902669 | 1.15 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr10_-_95415484 | 1.14 |
ENSMUST00000172070.1
ENSMUST00000150432.1 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_-_34287770 | 1.14 |
ENSMUST00000174751.1
ENSMUST00000040655.6 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr13_+_112464070 | 1.11 |
ENSMUST00000183663.1
ENSMUST00000184311.1 ENSMUST00000183886.1 |
Il6st
|
interleukin 6 signal transducer |
| chr5_+_92925400 | 1.11 |
ENSMUST00000172706.1
|
Shroom3
|
shroom family member 3 |
| chr7_-_99626936 | 1.07 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr11_-_23770953 | 1.06 |
ENSMUST00000102864.3
|
Rel
|
reticuloendotheliosis oncogene |
| chr3_+_153973436 | 0.97 |
ENSMUST00000089948.5
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr19_-_58455161 | 0.96 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr11_-_48994172 | 0.93 |
ENSMUST00000146439.1
|
Tgtp1
|
T cell specific GTPase 1 |
| chr10_-_95415283 | 0.90 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_-_35000848 | 0.87 |
ENSMUST00000166828.3
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5 |
| chr17_-_56005566 | 0.87 |
ENSMUST00000043785.6
|
Stap2
|
signal transducing adaptor family member 2 |
| chr11_+_101176041 | 0.86 |
ENSMUST00000103109.3
|
Cntnap1
|
contactin associated protein-like 1 |
| chr9_-_45009590 | 0.84 |
ENSMUST00000102832.1
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr19_-_58454435 | 0.83 |
ENSMUST00000169850.1
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr15_-_89410230 | 0.83 |
ENSMUST00000109314.2
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr17_-_45686899 | 0.81 |
ENSMUST00000156254.1
|
Tmem63b
|
transmembrane protein 63b |
| chr5_-_105293699 | 0.79 |
ENSMUST00000050011.8
|
Gbp6
|
guanylate binding protein 6 |
| chr17_-_28517509 | 0.75 |
ENSMUST00000114792.1
ENSMUST00000177939.1 |
Fkbp5
|
FK506 binding protein 5 |
| chr9_+_55326913 | 0.74 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr12_+_111166413 | 0.72 |
ENSMUST00000021706.4
|
Traf3
|
TNF receptor-associated factor 3 |
| chr10_-_19015347 | 0.71 |
ENSMUST00000019997.4
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_-_58454580 | 0.70 |
ENSMUST00000129100.1
ENSMUST00000123957.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr9_-_44735189 | 0.68 |
ENSMUST00000034611.8
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr11_+_87664549 | 0.68 |
ENSMUST00000121782.2
|
Rnf43
|
ring finger protein 43 |
| chr16_+_31422268 | 0.68 |
ENSMUST00000089759.2
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr12_+_111166536 | 0.66 |
ENSMUST00000060274.6
|
Traf3
|
TNF receptor-associated factor 3 |
| chr7_-_19629355 | 0.66 |
ENSMUST00000049912.8
ENSMUST00000094762.3 ENSMUST00000098754.4 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr16_-_45693658 | 0.65 |
ENSMUST00000114562.2
ENSMUST00000036617.7 |
Tmprss7
|
transmembrane serine protease 7 |
| chr5_-_92328068 | 0.65 |
ENSMUST00000113093.3
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr7_+_30421724 | 0.63 |
ENSMUST00000108176.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr10_+_111506286 | 0.63 |
ENSMUST00000164773.1
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr4_-_45012093 | 0.63 |
ENSMUST00000131991.1
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr7_+_30422389 | 0.63 |
ENSMUST00000108175.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr3_+_90080442 | 0.63 |
ENSMUST00000127955.1
|
Tpm3
|
tropomyosin 3, gamma |
| chr12_+_104214538 | 0.61 |
ENSMUST00000121337.1
ENSMUST00000167049.1 ENSMUST00000101080.1 |
Serpina3f
|
serine (or cysteine) peptidase inhibitor, clade A, member 3F |
| chr11_+_87664274 | 0.60 |
ENSMUST00000092800.5
|
Rnf43
|
ring finger protein 43 |
| chr9_-_14614949 | 0.60 |
ENSMUST00000013220.6
ENSMUST00000160770.1 |
Amotl1
|
angiomotin-like 1 |
| chr9_+_107906866 | 0.60 |
ENSMUST00000035203.7
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr3_+_142496924 | 0.59 |
ENSMUST00000090127.2
|
Gbp5
|
guanylate binding protein 5 |
| chr2_-_160313616 | 0.59 |
ENSMUST00000109475.2
|
Gm826
|
predicted gene 826 |
| chr11_+_100860326 | 0.58 |
ENSMUST00000138083.1
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr11_-_99493112 | 0.57 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr1_+_171250416 | 0.57 |
ENSMUST00000111315.1
ENSMUST00000006570.5 |
Adamts4
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 4 |
| chr7_-_126200413 | 0.56 |
ENSMUST00000163959.1
|
Xpo6
|
exportin 6 |
| chr2_-_167060417 | 0.56 |
ENSMUST00000155281.1
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr9_+_90162978 | 0.56 |
ENSMUST00000113060.1
|
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr17_-_34862473 | 0.55 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr9_+_90163057 | 0.55 |
ENSMUST00000113059.1
ENSMUST00000167122.1 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr18_-_4352944 | 0.55 |
ENSMUST00000025078.2
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_-_156839790 | 0.53 |
ENSMUST00000134838.1
ENSMUST00000137463.1 ENSMUST00000149275.2 |
Gm14230
|
predicted gene 14230 |
| chr14_-_45530118 | 0.53 |
ENSMUST00000045905.6
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr7_+_122671378 | 0.51 |
ENSMUST00000182563.1
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr9_-_44526397 | 0.51 |
ENSMUST00000062215.7
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr10_-_127751707 | 0.51 |
ENSMUST00000079692.5
|
Gpr182
|
G protein-coupled receptor 182 |
| chr16_+_32914094 | 0.51 |
ENSMUST00000023491.6
ENSMUST00000170899.1 ENSMUST00000171118.1 ENSMUST00000170201.1 ENSMUST00000165616.1 ENSMUST00000135193.2 |
Lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr9_+_124021965 | 0.49 |
ENSMUST00000039171.7
|
Ccr3
|
chemokine (C-C motif) receptor 3 |
| chr15_-_79164477 | 0.49 |
ENSMUST00000040019.4
|
Sox10
|
SRY-box containing gene 10 |
| chr4_-_96664112 | 0.48 |
ENSMUST00000030299.7
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr2_-_120314141 | 0.47 |
ENSMUST00000054651.7
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr5_-_105139539 | 0.46 |
ENSMUST00000100961.4
ENSMUST00000031235.6 ENSMUST00000100962.3 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr18_-_20059478 | 0.45 |
ENSMUST00000075214.2
ENSMUST00000039247.4 |
Dsc2
|
desmocollin 2 |
| chr1_+_171214013 | 0.45 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chrX_+_36112110 | 0.44 |
ENSMUST00000033418.7
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr7_+_28766747 | 0.44 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chr12_+_111166349 | 0.43 |
ENSMUST00000117269.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr7_-_44815658 | 0.43 |
ENSMUST00000107893.1
|
Atf5
|
activating transcription factor 5 |
| chr18_-_38929148 | 0.43 |
ENSMUST00000134864.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr15_-_89170688 | 0.43 |
ENSMUST00000060808.9
|
Plxnb2
|
plexin B2 |
| chr5_+_106609098 | 0.43 |
ENSMUST00000167618.1
|
Gm17304
|
predicted gene, 17304 |
| chr1_-_135258449 | 0.42 |
ENSMUST00000003135.7
|
Elf3
|
E74-like factor 3 |
| chr6_-_129275360 | 0.42 |
ENSMUST00000032259.3
|
Cd69
|
CD69 antigen |
| chr16_-_55838827 | 0.42 |
ENSMUST00000096026.2
ENSMUST00000036273.6 ENSMUST00000114457.1 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr2_-_6213033 | 0.42 |
ENSMUST00000042658.2
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr2_+_91259822 | 0.41 |
ENSMUST00000138470.1
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr4_+_131921771 | 0.41 |
ENSMUST00000094666.3
|
Tmem200b
|
transmembrane protein 200B |
| chr17_-_35702297 | 0.41 |
ENSMUST00000135078.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr4_+_85205417 | 0.41 |
ENSMUST00000030212.8
ENSMUST00000107189.1 ENSMUST00000107184.1 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr18_+_4994600 | 0.40 |
ENSMUST00000140448.1
|
Svil
|
supervillin |
| chr17_+_47688992 | 0.40 |
ENSMUST00000156118.1
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr8_-_84042540 | 0.40 |
ENSMUST00000005601.7
|
Il27ra
|
interleukin 27 receptor, alpha |
| chr14_+_33941021 | 0.39 |
ENSMUST00000100720.1
|
Gdf2
|
growth differentiation factor 2 |
| chr17_-_24205514 | 0.39 |
ENSMUST00000097376.3
ENSMUST00000168410.2 ENSMUST00000167791.2 ENSMUST00000040474.7 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr12_+_86678685 | 0.39 |
ENSMUST00000021681.3
|
Vash1
|
vasohibin 1 |
| chr2_+_144556306 | 0.39 |
ENSMUST00000155876.1
ENSMUST00000149697.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chr1_+_171213969 | 0.37 |
ENSMUST00000005820.4
ENSMUST00000075469.5 ENSMUST00000155126.1 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_18054258 | 0.37 |
ENSMUST00000026120.6
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr10_-_41709297 | 0.37 |
ENSMUST00000019955.9
ENSMUST00000099932.3 |
Ccdc162
|
coiled-coil domain containing 162 |
| chr2_-_5063996 | 0.37 |
ENSMUST00000114996.1
|
Optn
|
optineurin |
| chr18_+_37477768 | 0.37 |
ENSMUST00000051442.5
|
Pcdhb16
|
protocadherin beta 16 |
| chr2_-_160327494 | 0.37 |
ENSMUST00000099127.2
|
Gm826
|
predicted gene 826 |
| chr16_-_33056174 | 0.37 |
ENSMUST00000115100.1
ENSMUST00000040309.8 |
Iqcg
|
IQ motif containing G |
| chr7_+_96211656 | 0.36 |
ENSMUST00000107165.1
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr16_+_45093611 | 0.36 |
ENSMUST00000099498.2
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr9_-_70934808 | 0.36 |
ENSMUST00000034731.8
|
Lipc
|
lipase, hepatic |
| chr7_+_48959089 | 0.36 |
ENSMUST00000183659.1
|
Nav2
|
neuron navigator 2 |
| chr11_+_84129649 | 0.36 |
ENSMUST00000133811.1
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr14_+_53704007 | 0.36 |
ENSMUST00000103664.4
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr17_+_32685610 | 0.35 |
ENSMUST00000168171.1
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr17_+_32685655 | 0.35 |
ENSMUST00000008801.6
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr10_-_81430966 | 0.35 |
ENSMUST00000117966.1
|
Nfic
|
nuclear factor I/C |
| chr3_+_96601084 | 0.35 |
ENSMUST00000062058.3
|
Lix1l
|
Lix1-like |
| chr8_+_71676233 | 0.34 |
ENSMUST00000110013.3
ENSMUST00000051995.7 |
Jak3
|
Janus kinase 3 |
| chr15_+_99717515 | 0.34 |
ENSMUST00000023760.6
ENSMUST00000162194.1 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr17_+_35424842 | 0.33 |
ENSMUST00000174699.1
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr3_-_84305385 | 0.33 |
ENSMUST00000122849.1
ENSMUST00000132283.1 |
Trim2
|
tripartite motif-containing 2 |
| chr8_-_25101734 | 0.33 |
ENSMUST00000098866.4
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr18_+_36560581 | 0.33 |
ENSMUST00000155329.2
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr14_-_36968679 | 0.33 |
ENSMUST00000067700.6
|
Ccser2
|
coiled-coil serine rich 2 |
| chr7_-_31111148 | 0.33 |
ENSMUST00000164929.1
|
Hpn
|
hepsin |
| chr18_-_15403680 | 0.33 |
ENSMUST00000079081.6
|
Aqp4
|
aquaporin 4 |
| chr1_+_165302625 | 0.33 |
ENSMUST00000111450.1
|
Gpr161
|
G protein-coupled receptor 161 |
| chr4_+_102589687 | 0.32 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr14_-_70429072 | 0.32 |
ENSMUST00000048129.4
|
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr17_+_35379608 | 0.32 |
ENSMUST00000081435.4
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr10_+_4611971 | 0.32 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr11_+_100860447 | 0.32 |
ENSMUST00000107357.2
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr1_+_130800902 | 0.32 |
ENSMUST00000112477.2
ENSMUST00000027670.3 |
Fcamr
|
Fc receptor, IgA, IgM, high affinity |
| chr3_-_135691512 | 0.32 |
ENSMUST00000029812.7
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr1_+_88055467 | 0.31 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_77519186 | 0.31 |
ENSMUST00000100807.2
|
Gm10392
|
predicted gene 10392 |
| chr3_+_92316705 | 0.31 |
ENSMUST00000061038.2
|
Sprr2b
|
small proline-rich protein 2B |
| chr7_+_27029074 | 0.31 |
ENSMUST00000075552.5
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12 |
| chr15_-_76918010 | 0.31 |
ENSMUST00000048854.7
|
Zfp647
|
zinc finger protein 647 |
| chr6_-_124741374 | 0.31 |
ENSMUST00000004389.5
|
Grcc10
|
gene rich cluster, C10 gene |
| chr6_-_18030435 | 0.31 |
ENSMUST00000010941.2
|
Wnt2
|
wingless-related MMTV integration site 2 |
| chr11_+_100859492 | 0.30 |
ENSMUST00000107356.1
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr11_+_48838672 | 0.30 |
ENSMUST00000129674.1
|
Trim7
|
tripartite motif-containing 7 |
| chr16_+_32431225 | 0.30 |
ENSMUST00000115140.1
|
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha isoform |
| chr2_+_92185438 | 0.30 |
ENSMUST00000128781.2
|
Phf21a
|
PHD finger protein 21A |
| chr8_+_36457548 | 0.30 |
ENSMUST00000135373.1
ENSMUST00000147525.1 |
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr2_+_124089961 | 0.29 |
ENSMUST00000103241.1
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr10_+_13966268 | 0.29 |
ENSMUST00000015645.4
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr3_-_113258837 | 0.29 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr6_-_24956106 | 0.29 |
ENSMUST00000127247.2
|
Tmem229a
|
transmembrane protein 229A |
| chr7_-_25788635 | 0.29 |
ENSMUST00000002677.4
ENSMUST00000085948.4 |
Axl
|
AXL receptor tyrosine kinase |
| chr9_-_41157459 | 0.29 |
ENSMUST00000136530.1
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr11_+_49087022 | 0.29 |
ENSMUST00000046704.6
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr3_+_142620596 | 0.29 |
ENSMUST00000165774.1
|
Gbp2
|
guanylate binding protein 2 |
| chr7_+_75848338 | 0.29 |
ENSMUST00000092073.5
ENSMUST00000171155.2 |
Klhl25
|
kelch-like 25 |
| chr17_+_34187545 | 0.28 |
ENSMUST00000170086.1
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_-_35979679 | 0.28 |
ENSMUST00000173724.1
ENSMUST00000172900.1 ENSMUST00000174849.1 |
Prr3
|
proline-rich polypeptide 3 |
| chr10_-_87008015 | 0.28 |
ENSMUST00000035288.8
|
Stab2
|
stabilin 2 |
| chr17_-_45685973 | 0.28 |
ENSMUST00000145873.1
|
Tmem63b
|
transmembrane protein 63b |
| chr16_-_11176056 | 0.28 |
ENSMUST00000142389.1
ENSMUST00000138185.1 |
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr17_+_45555693 | 0.28 |
ENSMUST00000024742.7
|
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr13_+_33964659 | 0.28 |
ENSMUST00000021843.5
ENSMUST00000058978.7 |
Nqo2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr2_-_104028287 | 0.27 |
ENSMUST00000056170.3
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr6_+_129577893 | 0.27 |
ENSMUST00000053708.6
|
Klre1
|
killer cell lectin-like receptor family E member 1 |
| chr13_+_55152640 | 0.27 |
ENSMUST00000005452.5
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr9_+_65265173 | 0.27 |
ENSMUST00000048762.1
|
Cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr14_-_7994563 | 0.27 |
ENSMUST00000026315.7
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr17_-_45686120 | 0.27 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr8_+_71597648 | 0.27 |
ENSMUST00000143662.1
|
Fam129c
|
family with sequence similarity 129, member C |
| chr18_-_35722330 | 0.27 |
ENSMUST00000133064.1
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr10_+_128790903 | 0.27 |
ENSMUST00000026411.6
|
Mmp19
|
matrix metallopeptidase 19 |
| chr17_+_34187789 | 0.26 |
ENSMUST00000041633.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr9_-_7873157 | 0.26 |
ENSMUST00000159323.1
ENSMUST00000115673.2 |
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr3_-_83841767 | 0.26 |
ENSMUST00000029623.9
|
Tlr2
|
toll-like receptor 2 |
| chr10_+_25359798 | 0.26 |
ENSMUST00000053748.8
|
Epb4.1l2
|
erythrocyte protein band 4.1-like 2 |
| chr6_+_48841633 | 0.26 |
ENSMUST00000168406.1
|
Tmem176a
|
transmembrane protein 176A |
| chr14_-_77036641 | 0.26 |
ENSMUST00000062789.8
|
Lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr5_-_5663263 | 0.26 |
ENSMUST00000148193.1
|
A330021E22Rik
|
RIKEN cDNA A330021E22 gene |
| chr10_-_77089428 | 0.26 |
ENSMUST00000156009.1
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr2_-_5063932 | 0.26 |
ENSMUST00000027986.4
|
Optn
|
optineurin |
| chr6_+_48841476 | 0.26 |
ENSMUST00000101426.4
|
Tmem176a
|
transmembrane protein 176A |
| chr16_-_18235074 | 0.26 |
ENSMUST00000076957.5
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr1_+_88055377 | 0.26 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_164048214 | 0.25 |
ENSMUST00000027874.5
|
Sele
|
selectin, endothelial cell |
| chr14_-_103843685 | 0.25 |
ENSMUST00000172237.1
|
Ednrb
|
endothelin receptor type B |
| chr18_+_51117754 | 0.25 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr10_+_86705811 | 0.25 |
ENSMUST00000061458.7
ENSMUST00000075632.6 |
BC030307
|
cDNA sequence BC030307 |
| chr2_-_181202789 | 0.25 |
ENSMUST00000016511.5
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr17_+_8236031 | 0.25 |
ENSMUST00000164411.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr7_-_126200397 | 0.25 |
ENSMUST00000009344.9
|
Xpo6
|
exportin 6 |
| chr8_+_67490758 | 0.25 |
ENSMUST00000026677.3
|
Nat1
|
N-acetyl transferase 1 |
| chr4_+_42240639 | 0.24 |
ENSMUST00000117202.2
|
Gm10600
|
predicted gene 10600 |
| chr11_+_101552188 | 0.24 |
ENSMUST00000147239.1
|
Nbr1
|
neighbor of Brca1 gene 1 |
| chr7_+_101108768 | 0.24 |
ENSMUST00000098250.3
ENSMUST00000032931.7 |
Fchsd2
|
FCH and double SH3 domains 2 |
| chrX_-_100594860 | 0.24 |
ENSMUST00000053373.1
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr4_+_42091207 | 0.24 |
ENSMUST00000178882.1
|
Gm3893
|
predicted gene 3893 |
| chr1_-_36273425 | 0.24 |
ENSMUST00000056946.6
|
Neurl3
|
neuralized homolog 3 homolog (Drosophila) |
| chr19_-_5845471 | 0.24 |
ENSMUST00000174287.1
ENSMUST00000173672.1 |
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.5 | 1.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.4 | 1.6 | GO:2000503 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 1.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.1 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.3 | 3.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 1.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.3 | 1.2 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.3 | 1.3 | GO:1902623 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of neutrophil migration(GO:1902623) |
| 0.3 | 0.8 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.3 | 1.3 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 | 0.5 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.2 | 3.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 1.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 2.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.6 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 1.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 1.3 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.1 | 0.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.4 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.1 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:0000239 | pachytene(GO:0000239) |
| 0.1 | 0.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.3 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 1.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 2.0 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.3 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 0.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.4 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 1.7 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.5 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 1.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.5 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 2.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.1 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.1 | 0.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 1.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.7 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.1 | GO:0046271 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.3 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.3 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 0.2 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.3 | GO:0060750 | Sertoli cell proliferation(GO:0060011) prostate epithelial cord elongation(GO:0060523) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 1.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.3 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 1.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 1.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.4 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.5 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 1.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:2001182 | regulation of interleukin-12 secretion(GO:2001182) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0061235 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.2 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.0 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.0 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.0 | 0.5 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 1.1 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0021830 | substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.3 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.3 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0090650 | memory T cell differentiation(GO:0043379) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.0 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0035933 | glucocorticoid secretion(GO:0035933) regulation of glucocorticoid secretion(GO:2000849) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 3.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 2.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 3.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 8.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 4.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.5 | 1.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 5.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 2.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 1.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.2 | 3.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 1.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 3.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.4 | GO:0042903 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 4.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.2 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 1.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 2.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 2.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 8.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |