Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rest
Z-value: 0.89
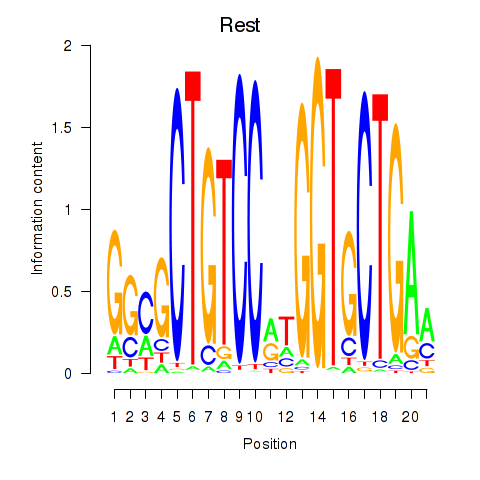
Transcription factors associated with Rest
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rest
|
ENSMUSG00000029249.9 | RE1-silencing transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rest | mm10_v2_chr5_+_77265454_77265549 | -0.21 | 2.1e-01 | Click! |
Activity profile of Rest motif
Sorted Z-values of Rest motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_23890805 | 13.61 |
ENSMUST00000004480.3
|
Sst
|
somatostatin |
| chr12_+_102554966 | 4.54 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr13_-_23991158 | 3.36 |
ENSMUST00000021770.7
|
Scgn
|
secretagogin, EF-hand calcium binding protein |
| chr2_+_132781278 | 2.46 |
ENSMUST00000028826.3
|
Chgb
|
chromogranin B |
| chr2_-_181314500 | 1.78 |
ENSMUST00000103045.3
|
Stmn3
|
stathmin-like 3 |
| chr7_+_27653906 | 1.76 |
ENSMUST00000008088.7
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr11_+_74619594 | 1.64 |
ENSMUST00000100866.2
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene |
| chrX_-_73869804 | 1.62 |
ENSMUST00000066576.5
ENSMUST00000114430.1 |
L1cam
|
L1 cell adhesion molecule |
| chr13_-_99900645 | 1.59 |
ENSMUST00000022150.6
|
Cartpt
|
CART prepropeptide |
| chr2_+_153031852 | 1.48 |
ENSMUST00000037235.6
|
Xkr7
|
X Kell blood group precursor related family member 7 homolog |
| chr10_+_5639210 | 1.40 |
ENSMUST00000019906.4
|
Vip
|
vasoactive intestinal polypeptide |
| chr15_-_76722161 | 1.20 |
ENSMUST00000049956.4
|
Lrrc24
|
leucine rich repeat containing 24 |
| chr5_+_137030275 | 1.15 |
ENSMUST00000041543.8
|
Vgf
|
VGF nerve growth factor inducible |
| chr7_-_81493883 | 1.13 |
ENSMUST00000082090.7
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr1_-_193035651 | 1.08 |
ENSMUST00000016344.7
|
Syt14
|
synaptotagmin XIV |
| chr19_+_42255704 | 1.05 |
ENSMUST00000087123.5
|
Golga7b
|
golgi autoantigen, golgin subfamily a, 7B |
| chr12_-_84698769 | 0.91 |
ENSMUST00000095550.2
|
Syndig1l
|
synapse differentiation inducing 1 like |
| chr11_+_77930800 | 0.84 |
ENSMUST00000093995.3
ENSMUST00000000646.7 |
Sez6
|
seizure related gene 6 |
| chr13_-_54749849 | 0.82 |
ENSMUST00000135343.1
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr1_-_84696182 | 0.75 |
ENSMUST00000049126.6
|
Dner
|
delta/notch-like EGF-related receptor |
| chr13_-_54749627 | 0.73 |
ENSMUST00000099506.1
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr15_-_75747922 | 0.70 |
ENSMUST00000062002.4
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chrX_+_7638674 | 0.66 |
ENSMUST00000128890.1
|
Syp
|
synaptophysin |
| chr2_-_27142429 | 0.53 |
ENSMUST00000151224.2
|
Fam163b
|
family with sequence similarity 163, member B |
| chr15_-_76521902 | 0.53 |
ENSMUST00000164703.1
ENSMUST00000096365.3 |
Scrt1
|
scratch homolog 1, zinc finger protein (Drosophila) |
| chr8_+_84946987 | 0.50 |
ENSMUST00000067472.7
ENSMUST00000109740.2 |
Rtbdn
|
retbindin |
| chr9_+_108826320 | 0.49 |
ENSMUST00000024238.5
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) |
| chr7_+_63444741 | 0.45 |
ENSMUST00000058476.7
|
Otud7a
|
OTU domain containing 7A |
| chr17_+_69439326 | 0.43 |
ENSMUST00000169935.1
|
A330050F15Rik
|
RIKEN cDNA A330050F15 gene |
| chr5_-_31138829 | 0.43 |
ENSMUST00000043475.2
|
Ucn
|
urocortin |
| chr7_-_79594924 | 0.43 |
ENSMUST00000172788.1
|
Rhcg
|
Rhesus blood group-associated C glycoprotein |
| chr11_+_69914179 | 0.41 |
ENSMUST00000057884.5
|
Gps2
|
G protein pathway suppressor 2 |
| chr15_+_101473472 | 0.41 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr12_-_112511136 | 0.40 |
ENSMUST00000066791.5
|
Tmem179
|
transmembrane protein 179 |
| chr6_-_121473630 | 0.37 |
ENSMUST00000046373.5
|
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr2_-_122313644 | 0.37 |
ENSMUST00000147788.1
ENSMUST00000154412.1 ENSMUST00000110537.1 ENSMUST00000148417.1 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr2_+_155775333 | 0.37 |
ENSMUST00000029141.5
|
Mmp24
|
matrix metallopeptidase 24 |
| chr7_+_19345135 | 0.33 |
ENSMUST00000160369.1
|
Ercc1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 |
| chr18_-_31447383 | 0.33 |
ENSMUST00000025110.3
|
Syt4
|
synaptotagmin IV |
| chr10_+_6788809 | 0.33 |
ENSMUST00000105611.1
ENSMUST00000052751.7 ENSMUST00000135502.1 ENSMUST00000105605.1 ENSMUST00000154906.1 ENSMUST00000092731.4 ENSMUST00000000783.6 ENSMUST00000078634.5 ENSMUST00000105602.1 ENSMUST00000105604.1 ENSMUST00000105603.1 |
Oprm1
|
opioid receptor, mu 1 |
| chr4_+_130055010 | 0.32 |
ENSMUST00000123617.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr2_-_25319095 | 0.32 |
ENSMUST00000114318.3
ENSMUST00000114310.3 ENSMUST00000114308.3 ENSMUST00000114317.3 ENSMUST00000028335.6 ENSMUST00000114314.3 ENSMUST00000114307.1 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr2_-_180104463 | 0.30 |
ENSMUST00000056480.3
|
Hrh3
|
histamine receptor H3 |
| chr1_+_70725902 | 0.29 |
ENSMUST00000161937.1
ENSMUST00000162182.1 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr17_-_83631892 | 0.27 |
ENSMUST00000051482.1
|
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr19_+_22139028 | 0.26 |
ENSMUST00000099569.2
ENSMUST00000087576.4 ENSMUST00000074770.5 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr9_-_63146980 | 0.26 |
ENSMUST00000055281.7
ENSMUST00000119146.1 |
Skor1
|
SKI family transcriptional corepressor 1 |
| chr1_+_70725715 | 0.26 |
ENSMUST00000053922.5
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr1_+_81077204 | 0.25 |
ENSMUST00000123720.1
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr2_+_152143552 | 0.23 |
ENSMUST00000089112.5
|
Tcf15
|
transcription factor 15 |
| chr11_-_35798884 | 0.22 |
ENSMUST00000160726.2
|
Fbll1
|
fibrillarin-like 1 |
| chr9_+_59578192 | 0.22 |
ENSMUST00000118549.1
ENSMUST00000034840.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr8_+_18846251 | 0.21 |
ENSMUST00000149565.1
ENSMUST00000033847.4 |
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr14_+_32321987 | 0.21 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr13_-_56895737 | 0.20 |
ENSMUST00000022023.6
ENSMUST00000109871.1 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr12_+_119945957 | 0.20 |
ENSMUST00000058644.8
|
Tmem196
|
transmembrane protein 196 |
| chr8_+_69088646 | 0.17 |
ENSMUST00000006435.7
|
Atp6v1b2
|
ATPase, H+ transporting, lysosomal V1 subunit B2 |
| chr8_-_104310032 | 0.17 |
ENSMUST00000160596.1
ENSMUST00000164175.1 |
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr2_-_25319187 | 0.16 |
ENSMUST00000114312.1
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr9_+_40873981 | 0.15 |
ENSMUST00000067375.3
|
Bsx
|
brain specific homeobox |
| chr7_-_27396542 | 0.15 |
ENSMUST00000108363.1
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr10_+_90829409 | 0.15 |
ENSMUST00000182202.1
ENSMUST00000182966.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_-_45046614 | 0.14 |
ENSMUST00000145391.1
|
Pdzd7
|
PDZ domain containing 7 |
| chr4_-_129121234 | 0.13 |
ENSMUST00000030572.3
|
Hpca
|
hippocalcin |
| chr2_-_28916668 | 0.13 |
ENSMUST00000113847.1
|
Barhl1
|
BarH-like 1 (Drosophila) |
| chr11_+_76202007 | 0.13 |
ENSMUST00000094014.3
|
Fam57a
|
family with sequence similarity 57, member A |
| chr8_-_104310109 | 0.13 |
ENSMUST00000159039.1
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr15_-_11037968 | 0.13 |
ENSMUST00000058007.5
|
Rxfp3
|
relaxin family peptide receptor 3 |
| chr10_-_41072279 | 0.13 |
ENSMUST00000061796.6
|
Gpr6
|
G protein-coupled receptor 6 |
| chr11_-_55607733 | 0.13 |
ENSMUST00000108853.1
ENSMUST00000075603.4 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr2_+_157456917 | 0.12 |
ENSMUST00000109529.1
|
Src
|
Rous sarcoma oncogene |
| chr9_+_40269430 | 0.11 |
ENSMUST00000171835.2
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr1_-_162548484 | 0.11 |
ENSMUST00000028017.9
|
Mettl13
|
methyltransferase like 13 |
| chr2_+_22622183 | 0.11 |
ENSMUST00000028123.3
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr13_+_54371340 | 0.10 |
ENSMUST00000026985.8
|
Cplx2
|
complexin 2 |
| chr11_-_3774706 | 0.10 |
ENSMUST00000155197.1
|
Osbp2
|
oxysterol binding protein 2 |
| chr10_+_90829538 | 0.08 |
ENSMUST00000179694.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_-_26208281 | 0.07 |
ENSMUST00000054099.9
|
Lhx3
|
LIM homeobox protein 3 |
| chr19_-_8929323 | 0.07 |
ENSMUST00000096242.3
|
Rom1
|
rod outer segment membrane protein 1 |
| chr4_+_15881255 | 0.07 |
ENSMUST00000029876.1
|
Calb1
|
calbindin 1 |
| chr17_+_28575718 | 0.07 |
ENSMUST00000080780.6
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr2_-_92401237 | 0.07 |
ENSMUST00000050312.2
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr1_+_34579693 | 0.06 |
ENSMUST00000052670.8
|
Amer3
|
APC membrane recruitment 3 |
| chr9_+_40269273 | 0.04 |
ENSMUST00000176185.1
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr8_+_55940453 | 0.04 |
ENSMUST00000000275.7
|
Glra3
|
glycine receptor, alpha 3 subunit |
| chr11_+_76202084 | 0.03 |
ENSMUST00000169560.1
|
Fam57a
|
family with sequence similarity 57, member A |
| chr12_+_102949450 | 0.03 |
ENSMUST00000179002.1
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr17_+_25240166 | 0.03 |
ENSMUST00000063574.6
|
Tsr3
|
TSR3 20S rRNA accumulation |
| chr11_-_55608189 | 0.02 |
ENSMUST00000102716.3
|
Glra1
|
glycine receptor, alpha 1 subunit |
| chr5_+_38260372 | 0.01 |
ENSMUST00000119047.1
|
Tmem128
|
transmembrane protein 128 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rest
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.5 | 1.4 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.4 | 1.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 0.7 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 1.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.5 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 1.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.3 | GO:0006296 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.3 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.1 | 0.4 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 1.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.5 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.8 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.4 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 13.2 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 3.7 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 1.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.8 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 17.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.9 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 18.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.5 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.3 | GO:0004985 | melanocortin receptor activity(GO:0004977) opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |