Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rfx5
Z-value: 0.68
Transcription factors associated with Rfx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx5
|
ENSMUSG00000005774.6 | regulatory factor X, 5 (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx5 | mm10_v2_chr3_+_94954075_94954226 | 0.01 | 9.3e-01 | Click! |
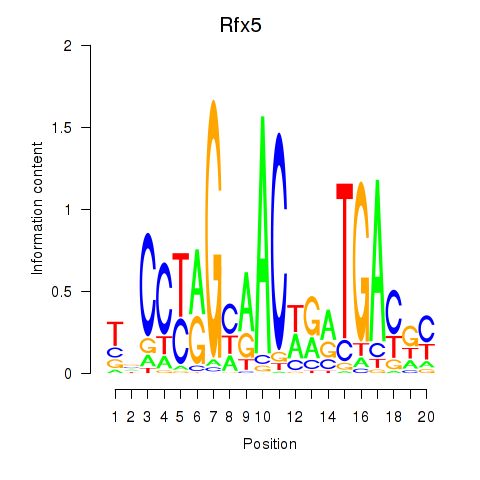
Activity profile of Rfx5 motif
Sorted Z-values of Rfx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_19692596 | 3.68 |
ENSMUST00000108451.2
ENSMUST00000045035.4 |
Apoc1
|
apolipoprotein C-I |
| chr5_-_145879857 | 2.83 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr17_+_34305883 | 2.42 |
ENSMUST00000074557.8
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta |
| chr17_+_34263209 | 2.26 |
ENSMUST00000040828.5
|
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr18_+_60803838 | 1.92 |
ENSMUST00000050487.8
ENSMUST00000097563.2 ENSMUST00000167610.1 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr1_+_88211956 | 1.90 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr17_+_34153072 | 1.69 |
ENSMUST00000114232.2
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr2_-_28563362 | 1.62 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr17_+_34187545 | 1.52 |
ENSMUST00000170086.1
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_-_34187219 | 1.49 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr7_+_101378183 | 1.44 |
ENSMUST00000084895.5
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_+_34197715 | 1.44 |
ENSMUST00000173441.1
ENSMUST00000025196.8 |
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr17_+_34187789 | 1.44 |
ENSMUST00000041633.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_+_122147680 | 1.38 |
ENSMUST00000102476.4
|
B2m
|
beta-2 microglobulin |
| chr17_+_35439155 | 1.36 |
ENSMUST00000071951.6
ENSMUST00000078205.7 ENSMUST00000116598.3 ENSMUST00000076256.7 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr12_+_108334341 | 1.31 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr17_+_35379608 | 1.29 |
ENSMUST00000081435.4
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr7_+_45277502 | 1.26 |
ENSMUST00000176276.1
|
1700039E15Rik
|
RIKEN cDNA 1700039E15 gene |
| chr14_+_53649387 | 1.06 |
ENSMUST00000103660.2
|
Trav15-2-dv6-2
|
T cell receptor alpha variable 15-2-DV6-2 |
| chr17_+_36121666 | 1.01 |
ENSMUST00000173128.1
|
Gm19684
|
predicted gene, 19684 |
| chr7_+_28180226 | 1.00 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr9_+_44772909 | 1.00 |
ENSMUST00000002099.3
|
Ift46
|
intraflagellar transport 46 |
| chr7_+_28180272 | 0.98 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr9_+_44772951 | 0.97 |
ENSMUST00000128150.1
|
Ift46
|
intraflagellar transport 46 |
| chr6_+_94500313 | 0.97 |
ENSMUST00000061118.8
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr8_+_27042555 | 0.96 |
ENSMUST00000033875.8
ENSMUST00000098851.4 |
Prosc
|
proline synthetase co-transcribed |
| chr17_+_34238914 | 0.90 |
ENSMUST00000167280.1
|
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr1_-_155146755 | 0.89 |
ENSMUST00000027744.8
|
Mr1
|
major histocompatibility complex, class I-related |
| chr5_-_125294107 | 0.83 |
ENSMUST00000127148.1
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr17_+_34238896 | 0.75 |
ENSMUST00000095342.3
|
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr9_+_44773191 | 0.75 |
ENSMUST00000147559.1
|
Ift46
|
intraflagellar transport 46 |
| chr5_+_104170676 | 0.70 |
ENSMUST00000112771.1
|
Dspp
|
dentin sialophosphoprotein |
| chr9_+_44773027 | 0.69 |
ENSMUST00000125877.1
|
Ift46
|
intraflagellar transport 46 |
| chr17_+_34135182 | 0.68 |
ENSMUST00000042121.9
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr7_+_28179469 | 0.67 |
ENSMUST00000085901.6
ENSMUST00000172761.1 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr12_-_87102350 | 0.65 |
ENSMUST00000110176.1
|
Ngb
|
neuroglobin |
| chr5_-_36830647 | 0.61 |
ENSMUST00000031002.3
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr19_+_32757497 | 0.60 |
ENSMUST00000013807.7
|
Pten
|
phosphatase and tensin homolog |
| chr17_+_37270214 | 0.59 |
ENSMUST00000038580.6
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr10_-_78307550 | 0.59 |
ENSMUST00000150828.1
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_-_101921186 | 0.54 |
ENSMUST00000106965.1
ENSMUST00000106968.1 ENSMUST00000106967.1 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr3_-_57692537 | 0.50 |
ENSMUST00000099091.3
|
Gm410
|
predicted gene 410 |
| chr17_-_24886304 | 0.46 |
ENSMUST00000044252.5
|
Nubp2
|
nucleotide binding protein 2 |
| chr17_+_34145231 | 0.44 |
ENSMUST00000171231.1
|
H2-DMb2
|
histocompatibility 2, class II, locus Mb2 |
| chr7_-_101921175 | 0.42 |
ENSMUST00000098236.2
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr17_+_34325658 | 0.39 |
ENSMUST00000050325.8
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr17_+_34145311 | 0.39 |
ENSMUST00000041982.7
|
H2-DMb2
|
histocompatibility 2, class II, locus Mb2 |
| chr5_+_11257157 | 0.34 |
ENSMUST00000177727.1
|
Gm8890
|
predicted gene 8890 |
| chr13_-_23488840 | 0.33 |
ENSMUST00000041541.4
ENSMUST00000110432.1 ENSMUST00000110433.3 |
Btn2a2
|
butyrophilin, subfamily 2, member A2 |
| chr1_+_194619815 | 0.33 |
ENSMUST00000027952.5
|
Plxna2
|
plexin A2 |
| chr4_+_107968332 | 0.32 |
ENSMUST00000106713.3
|
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr5_+_11128868 | 0.29 |
ENSMUST00000178863.1
|
Gm8879
|
predicted gene 8879 |
| chr5_+_11683236 | 0.29 |
ENSMUST00000178989.1
|
Gm8922
|
predicted gene 8922 |
| chr5_+_11184207 | 0.29 |
ENSMUST00000168407.2
|
Gm5861
|
predicted gene 5861 |
| chr3_-_95882031 | 0.28 |
ENSMUST00000161994.1
|
Gm129
|
predicted gene 129 |
| chr5_+_11342679 | 0.28 |
ENSMUST00000179482.1
|
Speer1-ps1
|
spermatogenesis associated glutamate (E)-rich protein 1, pseudogene 1 |
| chr5_+_11771686 | 0.28 |
ENSMUST00000178158.1
|
Gm8926
|
predicted gene 8926 |
| chr17_-_34305715 | 0.27 |
ENSMUST00000174074.1
|
Gm20513
|
predicted gene 20513 |
| chr11_+_117523526 | 0.26 |
ENSMUST00000132261.1
|
Gm11734
|
predicted gene 11734 |
| chr9_-_51102062 | 0.25 |
ENSMUST00000170947.1
|
4833427G06Rik
|
RIKEN cDNA 4833427G06 gene |
| chr5_+_10867186 | 0.25 |
ENSMUST00000168057.2
|
Gm6455
|
predicted gene 6455 |
| chr2_-_92434027 | 0.24 |
ENSMUST00000111278.1
ENSMUST00000090559.5 |
Cry2
|
cryptochrome 2 (photolyase-like) |
| chr7_-_45694369 | 0.24 |
ENSMUST00000040636.6
|
Sec1
|
secretory blood group 1 |
| chr17_+_24886643 | 0.23 |
ENSMUST00000117890.1
ENSMUST00000168265.1 ENSMUST00000120943.1 ENSMUST00000068508.6 ENSMUST00000119829.1 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr7_-_105752193 | 0.23 |
ENSMUST00000033184.4
|
Tpp1
|
tripeptidyl peptidase I |
| chr18_-_51865881 | 0.20 |
ENSMUST00000091905.2
|
Gm4950
|
predicted pseudogene 4950 |
| chr6_-_83831736 | 0.19 |
ENSMUST00000058383.8
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr14_+_41072936 | 0.16 |
ENSMUST00000161837.1
|
Dydc1
|
DPY30 domain containing 1 |
| chr3_-_95882193 | 0.15 |
ENSMUST00000159863.1
ENSMUST00000159739.1 ENSMUST00000036418.3 |
Gm129
|
predicted gene 129 |
| chr4_-_115781012 | 0.15 |
ENSMUST00000106521.1
|
Tex38
|
testis expressed 38 |
| chr14_+_111675113 | 0.14 |
ENSMUST00000042767.7
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr11_+_98682554 | 0.13 |
ENSMUST00000017365.8
|
Psmd3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr5_+_139150211 | 0.12 |
ENSMUST00000026975.6
|
Heatr2
|
HEAT repeat containing 2 |
| chr2_+_69380431 | 0.08 |
ENSMUST00000063690.3
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chrX_+_42526585 | 0.08 |
ENSMUST00000101619.3
|
Gm10483
|
predicted gene 10483 |
| chr3_+_16183177 | 0.08 |
ENSMUST00000108345.2
ENSMUST00000108346.2 |
Ythdf3
|
YTH domain family 3 |
| chr10_-_127263346 | 0.08 |
ENSMUST00000099172.3
|
Kif5a
|
kinesin family member 5A |
| chrX_+_45090882 | 0.07 |
ENSMUST00000040002.3
|
1110059M19Rik
|
RIKEN cDNA 1110059M19 gene |
| chr13_+_24831661 | 0.07 |
ENSMUST00000038039.2
|
Tdp2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr3_-_95882232 | 0.04 |
ENSMUST00000161866.1
|
Gm129
|
predicted gene 129 |
| chr10_-_7663569 | 0.01 |
ENSMUST00000162606.1
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.0 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.8 | 2.3 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.6 | 1.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.6 | 0.6 | GO:1901630 | negative regulation of presynaptic membrane organization(GO:1901630) |
| 0.6 | 3.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.6 | 1.7 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.5 | 7.5 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.3 | 0.8 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.2 | 2.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 0.7 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 2.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.3 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 3.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.2 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 2.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) adipose tissue development(GO:0060612) |
| 0.0 | 1.4 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 3.7 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 2.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 5.5 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.4 | 3.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 3.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.5 | 1.6 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.4 | 2.8 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.3 | 6.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 2.7 | GO:0046977 | TAP binding(GO:0046977) |
| 0.2 | 0.8 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 2.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 1.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 2.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 2.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 3.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |