Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
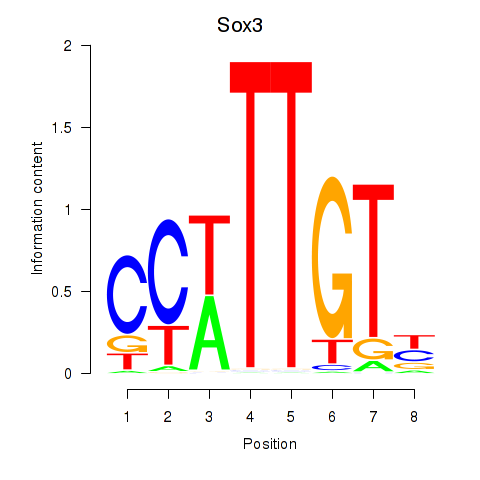
Results for Sox3_Sox10
Z-value: 0.63
Transcription factors associated with Sox3_Sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox3
|
ENSMUSG00000045179.8 | SRY (sex determining region Y)-box 3 |
|
Sox10
|
ENSMUSG00000033006.9 | SRY (sex determining region Y)-box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox10 | mm10_v2_chr15_-_79164477_79164496 | -0.01 | 9.6e-01 | Click! |
| Sox3 | mm10_v2_chrX_-_60893430_60893440 | 0.00 | 9.8e-01 | Click! |
Activity profile of Sox3_Sox10 motif
Sorted Z-values of Sox3_Sox10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_134915512 | 2.34 |
ENSMUST00000008987.4
|
Cldn13
|
claudin 13 |
| chr17_-_79355082 | 2.06 |
ENSMUST00000068958.7
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr10_+_43579161 | 1.97 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr4_+_115057683 | 1.94 |
ENSMUST00000161601.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr2_+_155611175 | 1.78 |
ENSMUST00000092995.5
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr7_-_100514800 | 1.43 |
ENSMUST00000054923.7
|
Dnajb13
|
DnaJ (Hsp40) related, subfamily B, member 13 |
| chr4_-_34882919 | 1.42 |
ENSMUST00000098163.2
ENSMUST00000047950.5 |
Zfp292
|
zinc finger protein 292 |
| chr4_-_148130678 | 1.35 |
ENSMUST00000030862.4
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr2_-_170406501 | 1.32 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr4_+_115057410 | 1.24 |
ENSMUST00000136946.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr11_+_67277124 | 1.17 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr15_-_36879816 | 1.16 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr11_-_96005872 | 1.14 |
ENSMUST00000013559.2
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr7_-_116038734 | 1.12 |
ENSMUST00000166877.1
|
Sox6
|
SRY-box containing gene 6 |
| chr14_+_31019183 | 1.07 |
ENSMUST00000052239.5
|
Pbrm1
|
polybromo 1 |
| chrX_+_93675088 | 1.06 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr6_+_86628174 | 1.05 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr9_-_103480328 | 1.04 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr12_+_109545390 | 1.04 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr14_+_80000292 | 1.03 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr14_-_70766598 | 1.00 |
ENSMUST00000167242.1
ENSMUST00000022696.6 |
Xpo7
|
exportin 7 |
| chr14_+_58075115 | 0.99 |
ENSMUST00000074654.5
|
Fgf9
|
fibroblast growth factor 9 |
| chr11_+_116531097 | 0.96 |
ENSMUST00000138840.1
|
Sphk1
|
sphingosine kinase 1 |
| chr7_-_120982260 | 0.94 |
ENSMUST00000033169.8
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr13_-_97747399 | 0.94 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr4_+_48585276 | 0.93 |
ENSMUST00000123476.1
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr6_+_47244359 | 0.90 |
ENSMUST00000060839.6
|
Cntnap2
|
contactin associated protein-like 2 |
| chr2_+_4559742 | 0.90 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr6_-_148944750 | 0.89 |
ENSMUST00000111562.1
ENSMUST00000081956.5 |
Fam60a
|
family with sequence similarity 60, member A |
| chr8_-_40634750 | 0.88 |
ENSMUST00000173957.1
|
Mtmr7
|
myotubularin related protein 7 |
| chr7_-_115824699 | 0.86 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr10_-_79874233 | 0.85 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chrX_+_93654863 | 0.85 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr4_-_58499398 | 0.85 |
ENSMUST00000107570.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr14_-_89898466 | 0.85 |
ENSMUST00000081204.4
|
Gm10110
|
predicted gene 10110 |
| chrX_+_157702574 | 0.84 |
ENSMUST00000112520.1
|
Smpx
|
small muscle protein, X-linked |
| chr9_+_112234257 | 0.83 |
ENSMUST00000149308.3
ENSMUST00000144424.2 ENSMUST00000139552.2 |
2900079G21Rik
|
RIKEN cDNA 2900079G21 gene |
| chr19_-_15924560 | 0.83 |
ENSMUST00000162053.1
|
Psat1
|
phosphoserine aminotransferase 1 |
| chrX_-_164258186 | 0.83 |
ENSMUST00000112265.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr4_+_107830958 | 0.82 |
ENSMUST00000106731.2
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr10_+_53596936 | 0.82 |
ENSMUST00000020004.6
|
Asf1a
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae) |
| chr16_+_17146937 | 0.82 |
ENSMUST00000115706.1
ENSMUST00000069064.4 |
Ydjc
|
YdjC homolog (bacterial) |
| chr18_+_24205937 | 0.80 |
ENSMUST00000164998.1
|
Galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr14_+_31019159 | 0.80 |
ENSMUST00000112094.1
ENSMUST00000144009.1 |
Pbrm1
|
polybromo 1 |
| chr6_-_52218686 | 0.79 |
ENSMUST00000134367.2
|
Hoxa7
|
homeobox A7 |
| chr18_+_82554463 | 0.79 |
ENSMUST00000062446.7
ENSMUST00000102812.4 ENSMUST00000075372.5 ENSMUST00000080658.4 ENSMUST00000152071.1 ENSMUST00000114674.3 ENSMUST00000142850.1 ENSMUST00000133193.1 ENSMUST00000123251.1 ENSMUST00000153478.1 ENSMUST00000132369.1 |
Mbp
|
myelin basic protein |
| chr4_+_48585135 | 0.79 |
ENSMUST00000030032.6
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr13_+_93304940 | 0.78 |
ENSMUST00000109497.1
ENSMUST00000109498.1 ENSMUST00000060490.4 ENSMUST00000109492.1 ENSMUST00000109496.1 ENSMUST00000109495.1 |
Homer1
|
homer homolog 1 (Drosophila) |
| chr2_-_152830615 | 0.77 |
ENSMUST00000146380.1
ENSMUST00000134902.1 ENSMUST00000134357.1 ENSMUST00000109820.3 |
Bcl2l1
|
BCL2-like 1 |
| chr14_-_65425453 | 0.76 |
ENSMUST00000059339.5
|
Pnoc
|
prepronociceptin |
| chrX_-_59568068 | 0.75 |
ENSMUST00000119833.1
ENSMUST00000131319.1 |
Fgf13
|
fibroblast growth factor 13 |
| chr5_+_26904682 | 0.75 |
ENSMUST00000120555.1
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr15_-_103251465 | 0.74 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr12_-_32208470 | 0.74 |
ENSMUST00000085469.5
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr13_-_97747373 | 0.72 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr12_+_109546409 | 0.71 |
ENSMUST00000143847.1
|
Meg3
|
maternally expressed 3 |
| chr14_+_31019125 | 0.70 |
ENSMUST00000112095.1
ENSMUST00000112098.3 ENSMUST00000112106.1 ENSMUST00000146325.1 |
Pbrm1
|
polybromo 1 |
| chr14_-_31019055 | 0.69 |
ENSMUST00000037739.6
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr4_-_132049058 | 0.69 |
ENSMUST00000105981.2
ENSMUST00000084253.3 ENSMUST00000141291.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr8_+_85432686 | 0.68 |
ENSMUST00000180883.1
|
1700051O22Rik
|
RIKEN cDNA 1700051O22 Gene |
| chr7_-_142095266 | 0.68 |
ENSMUST00000039926.3
|
Dusp8
|
dual specificity phosphatase 8 |
| chr12_-_32208609 | 0.68 |
ENSMUST00000053215.7
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr16_+_13986596 | 0.67 |
ENSMUST00000056521.5
ENSMUST00000118412.1 ENSMUST00000131608.1 |
2900011O08Rik
|
RIKEN cDNA 2900011O08 gene |
| chr14_+_79515618 | 0.67 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr2_-_153241402 | 0.67 |
ENSMUST00000056924.7
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr11_-_33843526 | 0.66 |
ENSMUST00000065970.5
ENSMUST00000109340.2 |
Kcnip1
|
Kv channel-interacting protein 1 |
| chr19_-_5894100 | 0.65 |
ENSMUST00000055911.4
|
Tigd3
|
tigger transposable element derived 3 |
| chr12_-_32061221 | 0.64 |
ENSMUST00000003079.5
ENSMUST00000036497.9 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr12_+_109546333 | 0.64 |
ENSMUST00000166636.2
|
Meg3
|
maternally expressed 3 |
| chr12_+_19606929 | 0.63 |
ENSMUST00000105167.1
|
Gm9257
|
predicted gene 9257 |
| chr11_-_100121558 | 0.63 |
ENSMUST00000007275.2
|
Krt13
|
keratin 13 |
| chrX_-_9256899 | 0.63 |
ENSMUST00000115553.2
|
Gm14862
|
predicted gene 14862 |
| chr3_+_103914560 | 0.62 |
ENSMUST00000106806.1
|
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr4_+_48585193 | 0.61 |
ENSMUST00000107703.1
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chrX_+_9272756 | 0.60 |
ENSMUST00000015486.6
|
Xk
|
Kell blood group precursor (McLeod phenotype) homolog |
| chr14_+_61138445 | 0.60 |
ENSMUST00000089394.3
ENSMUST00000119509.1 |
Sacs
|
sacsin |
| chr1_-_6215292 | 0.60 |
ENSMUST00000097832.1
|
4732440D04Rik
|
RIKEN cDNA 4732440D04 gene |
| chr8_-_85365341 | 0.59 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_+_123411424 | 0.59 |
ENSMUST00000071134.3
|
Tubb3
|
tubulin, beta 3 class III |
| chr10_-_30655859 | 0.58 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr1_-_97761538 | 0.58 |
ENSMUST00000171129.1
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr6_-_67037399 | 0.58 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr12_+_81631369 | 0.57 |
ENSMUST00000036116.5
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr14_+_14703025 | 0.57 |
ENSMUST00000057015.6
|
Slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr15_-_103255433 | 0.57 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr8_-_85365317 | 0.57 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr2_+_84734050 | 0.57 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chrX_+_109095359 | 0.57 |
ENSMUST00000033598.8
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chrX_+_166238901 | 0.56 |
ENSMUST00000112235.1
|
Gpm6b
|
glycoprotein m6b |
| chr13_+_93304799 | 0.55 |
ENSMUST00000080127.5
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr10_-_62792243 | 0.55 |
ENSMUST00000020268.5
|
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr12_-_54986363 | 0.55 |
ENSMUST00000173433.1
ENSMUST00000173803.1 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr13_+_117220584 | 0.55 |
ENSMUST00000022242.7
|
Emb
|
embigin |
| chr1_-_167466780 | 0.54 |
ENSMUST00000036643.4
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr12_+_17544873 | 0.54 |
ENSMUST00000171737.1
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr2_+_153492790 | 0.54 |
ENSMUST00000109783.1
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chrX_+_48519245 | 0.54 |
ENSMUST00000033430.2
|
Rab33a
|
RAB33A, member of RAS oncogene family |
| chr1_+_136131382 | 0.54 |
ENSMUST00000075164.4
|
Kif21b
|
kinesin family member 21B |
| chr1_-_66945019 | 0.54 |
ENSMUST00000027151.5
|
Myl1
|
myosin, light polypeptide 1 |
| chr8_+_123332676 | 0.54 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr5_+_66968559 | 0.54 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr2_+_173021902 | 0.53 |
ENSMUST00000029014.9
|
Rbm38
|
RNA binding motif protein 38 |
| chr7_-_113347273 | 0.53 |
ENSMUST00000117577.1
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chrX_+_166238923 | 0.53 |
ENSMUST00000060210.7
ENSMUST00000112233.1 |
Gpm6b
|
glycoprotein m6b |
| chr2_-_152831112 | 0.53 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr2_+_173022360 | 0.53 |
ENSMUST00000173997.1
|
Rbm38
|
RNA binding motif protein 38 |
| chr13_+_109685994 | 0.53 |
ENSMUST00000074103.5
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr12_-_36042476 | 0.52 |
ENSMUST00000020896.8
|
Tspan13
|
tetraspanin 13 |
| chr7_+_100493795 | 0.52 |
ENSMUST00000129324.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr13_+_75967704 | 0.52 |
ENSMUST00000022081.1
|
Spata9
|
spermatogenesis associated 9 |
| chr16_-_76373014 | 0.52 |
ENSMUST00000054178.1
|
Nrip1
|
nuclear receptor interacting protein 1 |
| chr14_+_27000362 | 0.52 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr17_-_45474839 | 0.52 |
ENSMUST00000024731.8
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr19_-_56822161 | 0.52 |
ENSMUST00000118592.1
|
A630007B06Rik
|
RIKEN cDNA A630007B06 gene |
| chr11_-_100135928 | 0.51 |
ENSMUST00000107411.2
|
Krt15
|
keratin 15 |
| chr6_+_65042575 | 0.51 |
ENSMUST00000031984.6
|
Smarcad1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_-_107794557 | 0.51 |
ENSMUST00000021066.3
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr8_+_57455898 | 0.51 |
ENSMUST00000034023.3
|
Scrg1
|
scrapie responsive gene 1 |
| chr7_-_4514558 | 0.51 |
ENSMUST00000163538.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr3_-_127837419 | 0.51 |
ENSMUST00000051737.6
|
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr7_-_4514609 | 0.50 |
ENSMUST00000166959.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr5_+_37245792 | 0.50 |
ENSMUST00000031004.7
|
Crmp1
|
collapsin response mediator protein 1 |
| chr2_-_126675224 | 0.50 |
ENSMUST00000124972.1
|
Gabpb1
|
GA repeat binding protein, beta 1 |
| chrX_-_47892502 | 0.50 |
ENSMUST00000077569.4
ENSMUST00000101616.2 ENSMUST00000088973.4 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr12_+_31438209 | 0.49 |
ENSMUST00000001254.5
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr17_+_3532554 | 0.49 |
ENSMUST00000168560.1
|
Cldn20
|
claudin 20 |
| chr2_-_131160006 | 0.48 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr11_+_97415527 | 0.48 |
ENSMUST00000121799.1
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr10_-_45470201 | 0.48 |
ENSMUST00000079390.6
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr19_+_55741810 | 0.47 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr3_-_96926020 | 0.47 |
ENSMUST00000062944.5
|
Gja8
|
gap junction protein, alpha 8 |
| chr6_-_82774448 | 0.46 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr6_+_86404336 | 0.46 |
ENSMUST00000113713.2
ENSMUST00000113708.1 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chrX_-_49788204 | 0.46 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr19_-_15924928 | 0.45 |
ENSMUST00000025542.3
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr14_+_55491062 | 0.45 |
ENSMUST00000076236.5
|
Lrrc16b
|
leucine rich repeat containing 16B |
| chr7_-_132813799 | 0.45 |
ENSMUST00000097998.2
|
Fam53b
|
family with sequence similarity 53, member B |
| chr12_+_51348265 | 0.45 |
ENSMUST00000119211.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr15_-_101694299 | 0.44 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr4_+_154960915 | 0.44 |
ENSMUST00000049621.6
|
Hes5
|
hairy and enhancer of split 5 (Drosophila) |
| chr6_-_86669136 | 0.44 |
ENSMUST00000001184.7
|
Mxd1
|
MAX dimerization protein 1 |
| chr2_-_168767136 | 0.44 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr11_+_9191934 | 0.44 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr10_+_17796256 | 0.43 |
ENSMUST00000037964.6
|
Txlnb
|
taxilin beta |
| chrX_-_59567348 | 0.43 |
ENSMUST00000124402.1
|
Fgf13
|
fibroblast growth factor 13 |
| chr12_+_51348370 | 0.43 |
ENSMUST00000121521.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr7_-_6331235 | 0.43 |
ENSMUST00000127658.1
ENSMUST00000062765.7 |
Zfp583
|
zinc finger protein 583 |
| chr16_+_92612755 | 0.43 |
ENSMUST00000180989.1
|
Gm26626
|
predicted gene, 26626 |
| chr6_-_87590701 | 0.42 |
ENSMUST00000050887.7
|
Prokr1
|
prokineticin receptor 1 |
| chr12_-_54986328 | 0.42 |
ENSMUST00000038926.6
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr17_-_68004075 | 0.42 |
ENSMUST00000024840.5
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr4_+_117849361 | 0.42 |
ENSMUST00000163288.1
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_163438455 | 0.42 |
ENSMUST00000109420.3
ENSMUST00000109421.3 ENSMUST00000018087.6 ENSMUST00000137070.1 |
Gdap1l1
|
ganglioside-induced differentiation-associated protein 1-like 1 |
| chr2_-_6884940 | 0.41 |
ENSMUST00000183091.1
ENSMUST00000182851.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr1_-_64122256 | 0.41 |
ENSMUST00000135075.1
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr16_+_87698904 | 0.41 |
ENSMUST00000026703.5
|
Bach1
|
BTB and CNC homology 1 |
| chr9_-_72111651 | 0.40 |
ENSMUST00000185117.1
|
Tcf12
|
transcription factor 12 |
| chr11_-_33843405 | 0.40 |
ENSMUST00000101368.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr4_+_13743424 | 0.40 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_-_95528702 | 0.40 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chrX_+_106920618 | 0.40 |
ENSMUST00000060576.7
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr1_-_136346074 | 0.40 |
ENSMUST00000048309.6
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr18_-_15063560 | 0.40 |
ENSMUST00000168989.1
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr17_+_53479212 | 0.39 |
ENSMUST00000017975.5
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr9_-_67832325 | 0.39 |
ENSMUST00000054500.5
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr2_-_6884975 | 0.39 |
ENSMUST00000114924.3
ENSMUST00000170438.1 ENSMUST00000114934.4 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr12_+_24831583 | 0.39 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chrX_-_47892396 | 0.39 |
ENSMUST00000153548.2
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr3_+_103914099 | 0.39 |
ENSMUST00000051139.6
ENSMUST00000068879.4 |
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr19_+_40831296 | 0.39 |
ENSMUST00000119316.1
|
Ccnj
|
cyclin J |
| chr4_-_43040279 | 0.38 |
ENSMUST00000107958.1
ENSMUST00000107959.1 ENSMUST00000152846.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr1_+_180641330 | 0.38 |
ENSMUST00000085804.5
|
Lin9
|
lin-9 homolog (C. elegans) |
| chr19_-_47692042 | 0.38 |
ENSMUST00000026045.7
ENSMUST00000086923.5 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr3_+_65109343 | 0.38 |
ENSMUST00000159525.1
ENSMUST00000049230.8 |
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr19_+_4214238 | 0.38 |
ENSMUST00000046506.6
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr2_+_131491958 | 0.37 |
ENSMUST00000110181.1
ENSMUST00000110180.1 |
Smox
|
spermine oxidase |
| chr10_-_82764088 | 0.37 |
ENSMUST00000130911.1
|
Nfyb
|
nuclear transcription factor-Y beta |
| chr7_-_115846080 | 0.37 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr6_+_86404257 | 0.37 |
ENSMUST00000095752.2
ENSMUST00000130967.1 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr5_+_74535449 | 0.37 |
ENSMUST00000080164.5
ENSMUST00000113536.1 ENSMUST00000122245.1 ENSMUST00000120618.1 ENSMUST00000113535.2 ENSMUST00000113534.2 |
Fip1l1
|
FIP1 like 1 (S. cerevisiae) |
| chr4_-_42874195 | 0.37 |
ENSMUST00000107978.1
ENSMUST00000055944.4 |
BC049635
|
cDNA sequence BC049635 |
| chr9_-_72111827 | 0.37 |
ENSMUST00000183404.1
ENSMUST00000184783.1 |
Tcf12
|
transcription factor 12 |
| chr1_-_174250976 | 0.36 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr13_-_3804307 | 0.36 |
ENSMUST00000077698.3
|
Calml3
|
calmodulin-like 3 |
| chr3_-_86920830 | 0.36 |
ENSMUST00000029719.8
|
Dclk2
|
doublecortin-like kinase 2 |
| chr2_-_168767029 | 0.36 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr2_-_169405435 | 0.36 |
ENSMUST00000131509.1
|
4930529I22Rik
|
RIKEN cDNA 4930529I22 gene |
| chrX_+_135993820 | 0.36 |
ENSMUST00000058119.7
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr12_+_24708984 | 0.36 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr18_+_11633276 | 0.36 |
ENSMUST00000115861.2
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr4_-_130574150 | 0.36 |
ENSMUST00000105993.3
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr14_+_26514554 | 0.36 |
ENSMUST00000104927.1
|
Gm2178
|
predicted gene 2178 |
| chr15_-_96642883 | 0.36 |
ENSMUST00000088452.4
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr4_+_6191093 | 0.35 |
ENSMUST00000029907.5
|
Ubxn2b
|
UBX domain protein 2B |
| chr2_+_131491764 | 0.35 |
ENSMUST00000028806.5
ENSMUST00000110179.2 ENSMUST00000110189.2 ENSMUST00000110182.2 ENSMUST00000110183.2 ENSMUST00000110186.2 ENSMUST00000110188.1 |
Smox
|
spermine oxidase |
| chr3_-_122619663 | 0.35 |
ENSMUST00000162409.1
|
Fnbp1l
|
formin binding protein 1-like |
| chr17_+_12119274 | 0.35 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr3_+_131110350 | 0.35 |
ENSMUST00000066849.6
ENSMUST00000106341.2 ENSMUST00000029611.7 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr3_-_84040151 | 0.35 |
ENSMUST00000052342.7
|
D930015E06Rik
|
RIKEN cDNA D930015E06 gene |
| chr13_-_75943812 | 0.35 |
ENSMUST00000022078.5
ENSMUST00000109606.1 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox3_Sox10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.7 | 2.1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.6 | 2.6 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.5 | 1.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.5 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 1.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 0.8 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.3 | 1.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 2.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.9 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 0.6 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 0.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 1.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.8 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.2 | 0.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 1.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 2.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 1.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 1.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.5 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.5 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 1.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.6 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 1.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 1.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.3 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.7 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 0.7 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.1 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.4 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 1.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.4 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 0.3 | GO:0006227 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.5 | GO:1904925 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 1.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 2.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.6 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.1 | 0.5 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.2 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.2 | GO:1904879 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) pentose-phosphate shunt, oxidative branch(GO:0009051) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.7 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.5 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.1 | 0.3 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.1 | GO:1902402 | signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.1 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 0.3 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.3 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.1 | GO:0061324 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) |
| 0.1 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.6 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.8 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.9 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.0 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 0.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.4 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 1.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.4 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 1.1 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.4 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.6 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 1.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.5 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.3 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 1.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.3 | GO:0061091 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) beta selection(GO:0043366) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 1.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.6 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.2 | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:0071839 | apoptotic process in bone marrow(GO:0071839) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.1 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.0 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0086070 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 1.1 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 2.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0021972 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance through spinal cord(GO:0021972) regulation of negative chemotaxis(GO:0050923) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.0 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.4 | 1.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 1.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 1.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 1.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.3 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 2.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.5 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.2 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.0 | 1.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 2.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.4 | 1.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.7 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.2 | 0.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 0.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.2 | 1.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 2.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.8 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.6 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 2.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.3 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.1 | 0.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.3 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.1 | 0.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 4.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 1.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 1.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 3.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |