Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
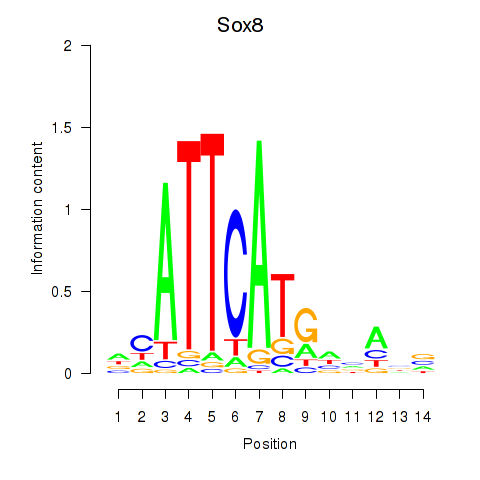
Results for Sox8
Z-value: 0.41
Transcription factors associated with Sox8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox8
|
ENSMUSG00000024176.4 | SRY (sex determining region Y)-box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox8 | mm10_v2_chr17_-_25570678_25570694 | 0.15 | 3.8e-01 | Click! |
Activity profile of Sox8 motif
Sorted Z-values of Sox8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87793470 | 1.77 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr11_+_32286946 | 1.68 |
ENSMUST00000101387.3
|
Hbq1b
|
hemoglobin, theta 1B |
| chr9_+_65101453 | 1.49 |
ENSMUST00000077696.6
ENSMUST00000035499.4 ENSMUST00000166273.1 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr13_+_65512678 | 1.35 |
ENSMUST00000081471.2
|
Gm10139
|
predicted gene 10139 |
| chr9_-_61976563 | 0.82 |
ENSMUST00000113990.1
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr11_-_26591729 | 0.71 |
ENSMUST00000109504.1
|
Vrk2
|
vaccinia related kinase 2 |
| chr7_-_126800354 | 0.57 |
ENSMUST00000106348.1
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr11_+_11487671 | 0.54 |
ENSMUST00000020410.4
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr7_-_126800036 | 0.53 |
ENSMUST00000133514.1
ENSMUST00000151137.1 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr8_-_33747724 | 0.48 |
ENSMUST00000179364.1
|
Smim18
|
small integral membrane protein 18 |
| chr8_+_94745590 | 0.46 |
ENSMUST00000034231.3
|
Ccl22
|
chemokine (C-C motif) ligand 22 |
| chr8_+_10006656 | 0.45 |
ENSMUST00000033892.7
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr17_-_22007301 | 0.44 |
ENSMUST00000075018.3
|
Gm9772
|
predicted gene 9772 |
| chr1_+_135783065 | 0.44 |
ENSMUST00000132795.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr12_-_8539545 | 0.42 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr2_+_4400958 | 0.41 |
ENSMUST00000075767.7
|
Frmd4a
|
FERM domain containing 4A |
| chr11_+_97415527 | 0.40 |
ENSMUST00000121799.1
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr4_+_11758147 | 0.39 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chr2_-_175131864 | 0.34 |
ENSMUST00000108929.2
|
Gm14399
|
predicted gene 14399 |
| chr7_-_42578588 | 0.29 |
ENSMUST00000179470.1
|
Gm21028
|
predicted gene, 21028 |
| chr15_-_103215285 | 0.27 |
ENSMUST00000122182.1
ENSMUST00000108813.3 ENSMUST00000127191.1 |
Cbx5
|
chromobox 5 |
| chr9_-_95858320 | 0.26 |
ENSMUST00000178038.1
|
1700065D16Rik
|
RIKEN cDNA 1700065D16 gene |
| chr6_-_72380416 | 0.26 |
ENSMUST00000101285.3
ENSMUST00000074231.3 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr5_+_17962621 | 0.24 |
ENSMUST00000030561.7
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr11_+_113659283 | 0.23 |
ENSMUST00000137878.1
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr14_-_49783327 | 0.22 |
ENSMUST00000118129.1
ENSMUST00000036972.6 |
3632451O06Rik
|
RIKEN cDNA 3632451O06 gene |
| chr9_-_32541589 | 0.21 |
ENSMUST00000016231.7
|
Fli1
|
Friend leukemia integration 1 |
| chrX_+_134404780 | 0.19 |
ENSMUST00000113224.2
ENSMUST00000113226.1 |
Drp2
|
dystrophin related protein 2 |
| chr11_-_99993992 | 0.19 |
ENSMUST00000105049.1
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr11_-_118909487 | 0.18 |
ENSMUST00000117731.1
ENSMUST00000106278.2 ENSMUST00000120061.1 ENSMUST00000017576.4 |
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr13_-_30950298 | 0.17 |
ENSMUST00000102946.1
|
Exoc2
|
exocyst complex component 2 |
| chr6_+_129397297 | 0.16 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr13_+_93304799 | 0.16 |
ENSMUST00000080127.5
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr15_-_99370427 | 0.16 |
ENSMUST00000081224.7
ENSMUST00000120633.1 ENSMUST00000088233.6 |
Fmnl3
|
formin-like 3 |
| chr13_-_66851513 | 0.16 |
ENSMUST00000169322.1
|
Gm17404
|
predicted gene, 17404 |
| chr3_-_92485886 | 0.16 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr3_-_8667033 | 0.14 |
ENSMUST00000042412.3
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1 |
| chr13_-_66852017 | 0.14 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr10_-_128180265 | 0.13 |
ENSMUST00000099139.1
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr11_-_43747963 | 0.12 |
ENSMUST00000048578.2
ENSMUST00000109278.1 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr18_-_62741387 | 0.12 |
ENSMUST00000097557.3
|
Spink13
|
serine peptidase inhibitor, Kazal type 13 |
| chr10_+_82378593 | 0.11 |
ENSMUST00000165906.1
|
Gm4924
|
predicted gene 4924 |
| chr4_-_134245579 | 0.11 |
ENSMUST00000030644.7
|
Zfp593
|
zinc finger protein 593 |
| chr6_+_92869357 | 0.11 |
ENSMUST00000113434.1
|
Gm15737
|
predicted gene 15737 |
| chr17_+_70561739 | 0.11 |
ENSMUST00000097288.2
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_-_69758630 | 0.10 |
ENSMUST00000058470.9
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr3_+_102010138 | 0.09 |
ENSMUST00000066187.4
|
Nhlh2
|
nescient helix loop helix 2 |
| chrX_-_72918284 | 0.08 |
ENSMUST00000152200.1
|
Cetn2
|
centrin 2 |
| chr18_+_36952621 | 0.08 |
ENSMUST00000115661.2
|
Pcdha2
|
protocadherin alpha 2 |
| chr4_-_58553553 | 0.07 |
ENSMUST00000107575.2
ENSMUST00000107574.1 ENSMUST00000147354.1 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr8_+_34807287 | 0.07 |
ENSMUST00000033930.4
|
Dusp4
|
dual specificity phosphatase 4 |
| chr7_+_43609907 | 0.06 |
ENSMUST00000116324.2
|
Zfp819
|
zinc finger protein 819 |
| chr4_-_129578535 | 0.05 |
ENSMUST00000052835.8
|
Fam167b
|
family with sequence similarity 167, member B |
| chr11_-_46166397 | 0.05 |
ENSMUST00000020679.2
|
Nipal4
|
NIPA-like domain containing 4 |
| chr4_-_116405986 | 0.04 |
ENSMUST00000123072.1
ENSMUST00000144281.1 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr1_+_109993982 | 0.04 |
ENSMUST00000027542.6
|
Cdh7
|
cadherin 7, type 2 |
| chr3_+_96104498 | 0.03 |
ENSMUST00000132980.1
ENSMUST00000138206.1 ENSMUST00000090785.2 ENSMUST00000035519.5 |
Otud7b
|
OTU domain containing 7B |
| chr9_-_95815389 | 0.02 |
ENSMUST00000119760.1
|
Pls1
|
plastin 1 (I-isoform) |
| chr1_+_109983737 | 0.02 |
ENSMUST00000172005.1
|
Cdh7
|
cadherin 7, type 2 |
| chrX_-_23285532 | 0.01 |
ENSMUST00000115319.2
|
Klhl13
|
kelch-like 13 |
| chr1_+_172341197 | 0.01 |
ENSMUST00000056136.3
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr4_+_154142363 | 0.00 |
ENSMUST00000030895.5
|
Wrap73
|
WD repeat containing, antisense to Trp73 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.5 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.7 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 1.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |