Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
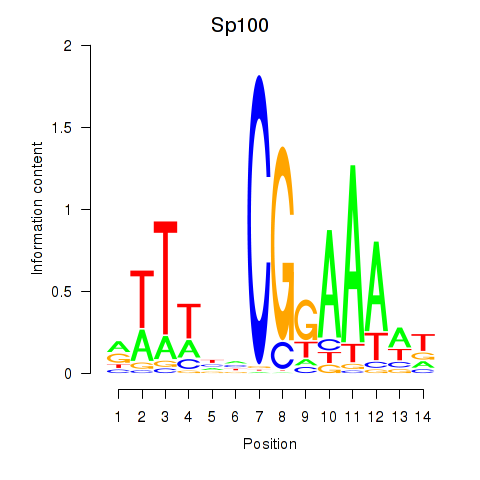
Results for Sp100
Z-value: 1.04
Transcription factors associated with Sp100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp100
|
ENSMUSG00000026222.10 | nuclear antigen Sp100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm10_v2_chr1_+_85650008_85650020 | -0.42 | 1.1e-02 | Click! |
Activity profile of Sp100 motif
Sorted Z-values of Sp100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_3023547 | 3.33 |
ENSMUST00000099046.3
|
Gm10718
|
predicted gene 10718 |
| chr2_-_98667264 | 3.27 |
ENSMUST00000099683.1
|
Gm10800
|
predicted gene 10800 |
| chr9_+_3013140 | 3.23 |
ENSMUST00000143083.2
|
Gm10721
|
predicted gene 10721 |
| chrX_-_61185558 | 2.98 |
ENSMUST00000166381.1
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr14_+_65806066 | 2.57 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr14_-_19418930 | 2.30 |
ENSMUST00000177817.1
|
Gm21738
|
predicted gene, 21738 |
| chr9_+_3000922 | 2.22 |
ENSMUST00000151376.2
|
Gm10722
|
predicted gene 10722 |
| chr9_+_3025417 | 2.13 |
ENSMUST00000075573.6
|
Gm10717
|
predicted gene 10717 |
| chr8_-_4779513 | 2.11 |
ENSMUST00000022945.7
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr9_+_3017408 | 2.10 |
ENSMUST00000099049.3
|
Gm10719
|
predicted gene 10719 |
| chr17_+_21691860 | 2.00 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr9_+_3015654 | 1.82 |
ENSMUST00000099050.3
|
Gm10720
|
predicted gene 10720 |
| chr9_+_3004457 | 1.64 |
ENSMUST00000178348.1
|
Gm11168
|
predicted gene 11168 |
| chr8_+_94179089 | 1.63 |
ENSMUST00000034215.6
|
Mt1
|
metallothionein 1 |
| chr12_-_74316394 | 1.57 |
ENSMUST00000110441.1
|
Gm11042
|
predicted gene 11042 |
| chr7_-_131327325 | 1.55 |
ENSMUST00000033146.7
|
1700007K09Rik
|
RIKEN cDNA 1700007K09 gene |
| chr9_+_3018753 | 1.52 |
ENSMUST00000179272.1
|
Gm10719
|
predicted gene 10719 |
| chr3_+_135212557 | 1.41 |
ENSMUST00000062893.7
|
Cenpe
|
centromere protein E |
| chr9_+_3027439 | 1.40 |
ENSMUST00000177875.1
ENSMUST00000179982.1 |
Gm10717
|
predicted gene 10717 |
| chr9_-_36726374 | 1.39 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr4_-_118809814 | 1.38 |
ENSMUST00000105035.1
ENSMUST00000084313.3 |
Olfr1335
|
olfactory receptor 1335 |
| chr6_+_18848571 | 1.25 |
ENSMUST00000056398.8
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr17_-_36190121 | 1.24 |
ENSMUST00000097329.3
ENSMUST00000025312.6 ENSMUST00000102675.3 |
H2-T3
|
histocompatibility 2, T region locus 3 |
| chr2_-_129297205 | 1.20 |
ENSMUST00000052708.6
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr9_+_3036877 | 1.10 |
ENSMUST00000155807.2
|
Gm10715
|
predicted gene 10715 |
| chr11_-_11808923 | 1.09 |
ENSMUST00000109664.1
ENSMUST00000150714.1 ENSMUST00000047689.4 ENSMUST00000171938.1 ENSMUST00000171080.1 |
Fignl1
|
fidgetin-like 1 |
| chr5_-_21785115 | 1.03 |
ENSMUST00000115193.1
ENSMUST00000115192.1 ENSMUST00000115195.1 ENSMUST00000030771.5 |
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr7_+_13733502 | 1.02 |
ENSMUST00000086148.6
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr7_+_13623967 | 0.96 |
ENSMUST00000108525.2
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr14_-_67715585 | 0.96 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr9_+_25089422 | 0.94 |
ENSMUST00000086238.2
|
Gm10181
|
predicted gene 10181 |
| chr14_-_87141206 | 0.94 |
ENSMUST00000022599.7
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr2_-_119618455 | 0.91 |
ENSMUST00000123818.1
|
Oip5
|
Opa interacting protein 5 |
| chr2_-_132578244 | 0.90 |
ENSMUST00000110142.1
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr17_-_74323896 | 0.85 |
ENSMUST00000164832.1
|
Dpy30
|
dpy-30 homolog (C. elegans) |
| chr5_-_86906937 | 0.85 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr4_-_116821501 | 0.81 |
ENSMUST00000055436.3
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr19_+_38931008 | 0.80 |
ENSMUST00000145051.1
|
Hells
|
helicase, lymphoid specific |
| chr3_+_122274371 | 0.80 |
ENSMUST00000035776.8
|
Dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr16_-_56029696 | 0.79 |
ENSMUST00000122253.1
ENSMUST00000114444.2 |
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr2_-_140170528 | 0.76 |
ENSMUST00000046030.7
|
Esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr14_-_87141114 | 0.73 |
ENSMUST00000168889.1
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr5_-_149053038 | 0.73 |
ENSMUST00000085546.6
|
Hmgb1
|
high mobility group box 1 |
| chr1_-_93342734 | 0.72 |
ENSMUST00000027493.3
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr8_+_70673364 | 0.71 |
ENSMUST00000146972.1
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_-_106003549 | 0.69 |
ENSMUST00000102555.4
|
Dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr7_-_13989588 | 0.69 |
ENSMUST00000165167.1
ENSMUST00000108520.2 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr15_+_85859689 | 0.69 |
ENSMUST00000170629.1
|
Gtse1
|
G two S phase expressed protein 1 |
| chr5_-_129787175 | 0.68 |
ENSMUST00000031399.6
|
Psph
|
phosphoserine phosphatase |
| chrX_-_104671048 | 0.68 |
ENSMUST00000042070.5
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr8_+_22411340 | 0.64 |
ENSMUST00000033934.3
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr18_+_34624621 | 0.64 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr2_+_136891501 | 0.63 |
ENSMUST00000141463.1
|
Slx4ip
|
SLX4 interacting protein |
| chr19_+_38930909 | 0.62 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr11_-_16508069 | 0.60 |
ENSMUST00000109641.1
|
Sec61g
|
SEC61, gamma subunit |
| chr12_-_81421910 | 0.60 |
ENSMUST00000085319.3
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr6_-_71908736 | 0.59 |
ENSMUST00000082094.2
|
Ptcd3
|
pentatricopeptide repeat domain 3 |
| chr11_-_116024489 | 0.59 |
ENSMUST00000016703.7
|
H3f3b
|
H3 histone, family 3B |
| chr4_+_21879662 | 0.59 |
ENSMUST00000029909.2
|
Coq3
|
coenzyme Q3 homolog, methyltransferase (yeast) |
| chr10_-_81202037 | 0.59 |
ENSMUST00000005069.6
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr8_-_4217261 | 0.58 |
ENSMUST00000168386.2
|
BC068157
|
cDNA sequence BC068157 |
| chrX_+_37126777 | 0.57 |
ENSMUST00000016553.4
|
Nkap
|
NFKB activating protein |
| chr6_-_87672142 | 0.57 |
ENSMUST00000032130.2
ENSMUST00000065997.2 |
Aplf
|
aprataxin and PNKP like factor |
| chr11_-_16508149 | 0.56 |
ENSMUST00000109642.1
|
Sec61g
|
SEC61, gamma subunit |
| chr5_-_87254804 | 0.56 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr9_+_109832749 | 0.54 |
ENSMUST00000147777.1
ENSMUST00000035053.5 ENSMUST00000133483.1 |
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr13_+_23575753 | 0.52 |
ENSMUST00000105105.1
|
Hist1h3d
|
histone cluster 1, H3d |
| chr12_-_69357120 | 0.51 |
ENSMUST00000021368.8
|
Nemf
|
nuclear export mediator factor |
| chr5_-_138272786 | 0.50 |
ENSMUST00000161279.1
ENSMUST00000161647.1 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr17_-_10320229 | 0.50 |
ENSMUST00000053066.6
|
Qk
|
quaking |
| chr4_+_105789869 | 0.50 |
ENSMUST00000184254.1
|
Gm12728
|
predicted gene 12728 |
| chr6_+_134981998 | 0.50 |
ENSMUST00000167323.1
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr13_-_22041352 | 0.49 |
ENSMUST00000102977.2
|
Hist1h4i
|
histone cluster 1, H4i |
| chr2_-_74578875 | 0.49 |
ENSMUST00000134168.1
ENSMUST00000111993.2 ENSMUST00000064503.6 |
Lnp
|
limb and neural patterns |
| chr17_+_47505149 | 0.48 |
ENSMUST00000183177.1
ENSMUST00000182848.1 |
Ccnd3
|
cyclin D3 |
| chr3_-_88410295 | 0.48 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chr12_+_3891728 | 0.48 |
ENSMUST00000172689.1
ENSMUST00000111186.1 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr3_-_158036630 | 0.48 |
ENSMUST00000121326.1
|
Srsf11
|
serine/arginine-rich splicing factor 11 |
| chr3_+_92967059 | 0.47 |
ENSMUST00000098886.4
|
Lce3e
|
late cornified envelope 3E |
| chr10_+_111125851 | 0.47 |
ENSMUST00000171120.1
|
Gm5428
|
predicted gene 5428 |
| chr9_+_109832998 | 0.46 |
ENSMUST00000119376.1
ENSMUST00000122343.1 |
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr14_-_33978751 | 0.46 |
ENSMUST00000166737.1
|
Zfp488
|
zinc finger protein 488 |
| chr13_+_74406387 | 0.46 |
ENSMUST00000090860.6
|
Gm10116
|
predicted pseudogene 10116 |
| chr13_+_23746734 | 0.46 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chrX_-_56822308 | 0.46 |
ENSMUST00000135542.1
ENSMUST00000114766.1 |
Mtap7d3
|
MAP7 domain containing 3 |
| chr14_+_75845296 | 0.45 |
ENSMUST00000142061.1
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr4_-_129623870 | 0.44 |
ENSMUST00000106035.1
ENSMUST00000150357.1 ENSMUST00000030586.8 |
Ccdc28b
|
coiled coil domain containing 28B |
| chr2_+_75659253 | 0.44 |
ENSMUST00000111964.1
ENSMUST00000111962.1 ENSMUST00000111961.1 ENSMUST00000164947.2 ENSMUST00000090792.4 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chrX_-_167209149 | 0.44 |
ENSMUST00000112176.1
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr4_+_21727726 | 0.44 |
ENSMUST00000102997.1
ENSMUST00000120679.1 ENSMUST00000108240.2 |
Ccnc
|
cyclin C |
| chr9_-_83254460 | 0.43 |
ENSMUST00000184080.1
ENSMUST00000184100.1 |
RP23-341H6.1
|
RP23-341H6.1 |
| chr15_+_77477044 | 0.43 |
ENSMUST00000060551.2
ENSMUST00000119997.1 |
Apol10a
|
apolipoprotein L 10A |
| chr17_+_22689771 | 0.43 |
ENSMUST00000055305.8
|
Gm9805
|
predicted gene 9805 |
| chr2_+_136892168 | 0.43 |
ENSMUST00000099311.2
|
Slx4ip
|
SLX4 interacting protein |
| chr6_+_96113146 | 0.43 |
ENSMUST00000122120.1
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chrX_-_60403947 | 0.43 |
ENSMUST00000033480.6
ENSMUST00000101527.2 |
Atp11c
|
ATPase, class VI, type 11C |
| chr2_+_105904629 | 0.42 |
ENSMUST00000037499.5
|
Immp1l
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_-_14123042 | 0.42 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr7_-_84679346 | 0.41 |
ENSMUST00000069537.2
ENSMUST00000178385.1 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr4_+_119195353 | 0.41 |
ENSMUST00000106345.2
|
Ccdc23
|
coiled-coil domain containing 23 |
| chr8_+_71922810 | 0.41 |
ENSMUST00000119003.1
|
Zfp617
|
zinc finger protein 617 |
| chr5_+_21785253 | 0.41 |
ENSMUST00000030769.5
|
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chr4_-_129619069 | 0.41 |
ENSMUST00000121442.1
ENSMUST00000046675.5 |
Iqcc
|
IQ motif containing C |
| chrX_+_74424534 | 0.40 |
ENSMUST00000135165.1
ENSMUST00000114128.1 ENSMUST00000114133.2 ENSMUST00000004330.3 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr2_+_62664279 | 0.40 |
ENSMUST00000028257.2
|
Gca
|
grancalcin |
| chr16_-_14317319 | 0.40 |
ENSMUST00000120707.1
ENSMUST00000023357.7 |
Fopnl
|
Fgfr1op N-terminal like |
| chr13_-_108158584 | 0.40 |
ENSMUST00000163558.1
|
Ndufaf2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2 |
| chr4_+_40948401 | 0.39 |
ENSMUST00000030128.5
|
Chmp5
|
charged multivesicular body protein 5 |
| chr5_-_124327883 | 0.39 |
ENSMUST00000031344.6
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr4_+_134923589 | 0.39 |
ENSMUST00000078084.6
|
D4Wsu53e
|
DNA segment, Chr 4, Wayne State University 53, expressed |
| chr18_-_64516547 | 0.39 |
ENSMUST00000025483.9
|
Nars
|
asparaginyl-tRNA synthetase |
| chr15_+_27025386 | 0.38 |
ENSMUST00000169678.2
|
Gm6576
|
predicted gene 6576 |
| chr9_+_54286479 | 0.38 |
ENSMUST00000056740.5
|
Gldn
|
gliomedin |
| chr10_-_86705485 | 0.38 |
ENSMUST00000020238.7
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr2_+_164316730 | 0.38 |
ENSMUST00000017144.2
|
Svs6
|
seminal vesicle secretory protein 6 |
| chr9_+_60794468 | 0.38 |
ENSMUST00000050183.6
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chrX_-_166510424 | 0.37 |
ENSMUST00000112188.1
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr1_+_107422681 | 0.37 |
ENSMUST00000112710.1
ENSMUST00000086690.4 |
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr4_-_14621805 | 0.37 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_-_118458747 | 0.37 |
ENSMUST00000097615.1
|
2900060B14Rik
|
RIKEN cDNA 2900060B14 gene |
| chr19_+_48206025 | 0.37 |
ENSMUST00000078880.5
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr2_-_127208274 | 0.36 |
ENSMUST00000056146.1
|
1810024B03Rik
|
RIKEN cDNA 1810024B03 gene |
| chr19_+_46689902 | 0.36 |
ENSMUST00000074912.7
|
2010012O05Rik
|
RIKEN cDNA 2010012O05 gene |
| chr1_-_65179058 | 0.36 |
ENSMUST00000097709.4
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr2_-_51934644 | 0.35 |
ENSMUST00000165313.1
|
Rbm43
|
RNA binding motif protein 43 |
| chr7_+_104003259 | 0.34 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr7_-_132599637 | 0.34 |
ENSMUST00000054562.3
|
Nkx1-2
|
NK1 transcription factor related, locus 2 (Drosophila) |
| chr2_+_166792525 | 0.34 |
ENSMUST00000065753.1
|
Trp53rk
|
transformation related protein 53 regulating kinase |
| chr2_-_136891363 | 0.34 |
ENSMUST00000028730.6
ENSMUST00000110089.2 |
Mkks
|
McKusick-Kaufman syndrome |
| chr7_-_5413145 | 0.34 |
ENSMUST00000108569.2
|
Vmn1r58
|
vomeronasal 1 receptor 58 |
| chr5_+_52834009 | 0.34 |
ENSMUST00000031072.7
|
Anapc4
|
anaphase promoting complex subunit 4 |
| chr12_+_98771018 | 0.34 |
ENSMUST00000021399.7
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr4_-_25281801 | 0.33 |
ENSMUST00000102994.3
|
Ufl1
|
UFM1 specific ligase 1 |
| chr7_-_133776772 | 0.33 |
ENSMUST00000033290.5
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_+_155803521 | 0.32 |
ENSMUST00000030942.6
ENSMUST00000185148.1 ENSMUST00000130188.1 |
Mrpl20
|
mitochondrial ribosomal protein L20 |
| chr2_+_29124106 | 0.32 |
ENSMUST00000129544.1
|
Setx
|
senataxin |
| chr13_+_49653297 | 0.32 |
ENSMUST00000021824.7
|
Nol8
|
nucleolar protein 8 |
| chr14_+_50807915 | 0.32 |
ENSMUST00000036126.5
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr12_+_9029982 | 0.32 |
ENSMUST00000085741.1
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr5_+_129787390 | 0.31 |
ENSMUST00000031402.8
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr6_-_23132981 | 0.31 |
ENSMUST00000031707.7
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr10_+_80249106 | 0.31 |
ENSMUST00000105364.1
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chr16_+_33380765 | 0.31 |
ENSMUST00000165418.1
|
Zfp148
|
zinc finger protein 148 |
| chr9_-_44440868 | 0.31 |
ENSMUST00000098837.1
|
Foxr1
|
forkhead box R1 |
| chr2_+_104027823 | 0.31 |
ENSMUST00000111135.1
ENSMUST00000111136.1 ENSMUST00000102565.3 |
Fbxo3
|
F-box protein 3 |
| chr2_-_25224653 | 0.30 |
ENSMUST00000043584.4
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr13_+_17695409 | 0.30 |
ENSMUST00000049744.3
|
Mplkip
|
M-phase specific PLK1 intereacting protein |
| chr6_+_117841174 | 0.30 |
ENSMUST00000112859.1
ENSMUST00000137224.1 ENSMUST00000164472.1 ENSMUST00000112861.1 ENSMUST00000035638.8 |
Zfp637
|
zinc finger protein 637 |
| chr19_-_34475135 | 0.30 |
ENSMUST00000050562.4
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr1_-_34439672 | 0.30 |
ENSMUST00000042493.8
|
Ccdc115
|
coiled-coil domain containing 115 |
| chr17_+_33524170 | 0.29 |
ENSMUST00000087623.6
|
Adamts10
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
| chr7_-_46710642 | 0.29 |
ENSMUST00000143082.1
|
Saal1
|
serum amyloid A-like 1 |
| chr10_-_128891674 | 0.29 |
ENSMUST00000026408.6
|
Gdf11
|
growth differentiation factor 11 |
| chr5_-_124352233 | 0.29 |
ENSMUST00000111472.1
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr8_-_56550791 | 0.28 |
ENSMUST00000134162.1
ENSMUST00000140107.1 ENSMUST00000040330.8 ENSMUST00000135337.1 |
Cep44
|
centrosomal protein 44 |
| chr6_+_52713729 | 0.28 |
ENSMUST00000080723.4
ENSMUST00000149588.1 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr8_-_11678728 | 0.28 |
ENSMUST00000033906.4
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr16_+_10411928 | 0.28 |
ENSMUST00000023146.4
|
Nubp1
|
nucleotide binding protein 1 |
| chr1_+_134709293 | 0.28 |
ENSMUST00000121990.1
|
Syt2
|
synaptotagmin II |
| chr14_+_56668242 | 0.28 |
ENSMUST00000116468.1
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr15_+_44196135 | 0.28 |
ENSMUST00000038856.6
ENSMUST00000110289.3 |
Trhr
|
thyrotropin releasing hormone receptor |
| chr14_-_103099499 | 0.27 |
ENSMUST00000022720.8
|
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr16_+_65520503 | 0.27 |
ENSMUST00000176330.1
ENSMUST00000004964.8 ENSMUST00000176038.1 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr9_-_59750616 | 0.27 |
ENSMUST00000163586.1
ENSMUST00000177963.1 ENSMUST00000051039.4 |
Senp8
|
SUMO/sentrin specific peptidase 8 |
| chr2_+_71055731 | 0.27 |
ENSMUST00000154704.1
ENSMUST00000135357.1 ENSMUST00000064141.5 ENSMUST00000112159.2 ENSMUST00000102701.3 |
Dcaf17
|
DDB1 and CUL4 associated factor 17 |
| chr11_+_78176711 | 0.27 |
ENSMUST00000098545.5
|
Tlcd1
|
TLC domain containing 1 |
| chr13_-_67500444 | 0.27 |
ENSMUST00000163534.1
ENSMUST00000091523.2 ENSMUST00000171518.1 ENSMUST00000076123.5 |
Zfp58
|
zinc finger protein 58 |
| chr4_-_40948196 | 0.27 |
ENSMUST00000030125.4
ENSMUST00000108089.1 |
Bag1
|
BCL2-associated athanogene 1 |
| chr6_+_18170687 | 0.27 |
ENSMUST00000045706.5
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr12_+_73964481 | 0.27 |
ENSMUST00000021532.4
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr2_-_136891131 | 0.26 |
ENSMUST00000144275.1
|
Mkks
|
McKusick-Kaufman syndrome |
| chr19_-_27429807 | 0.26 |
ENSMUST00000076219.4
|
D19Bwg1357e
|
DNA segment, Chr 19, Brigham & Women's Genetics 1357 expressed |
| chr6_-_112696604 | 0.26 |
ENSMUST00000113182.1
ENSMUST00000113180.1 ENSMUST00000068487.5 ENSMUST00000077088.4 |
Rad18
|
RAD18 homolog (S. cerevisiae) |
| chr11_-_115276973 | 0.26 |
ENSMUST00000021078.2
|
Fdxr
|
ferredoxin reductase |
| chr11_+_116198853 | 0.26 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr16_+_18836573 | 0.26 |
ENSMUST00000055413.6
|
2510002D24Rik
|
RIKEN cDNA 2510002D24 gene |
| chr10_+_80249441 | 0.26 |
ENSMUST00000020361.6
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chr17_+_47505211 | 0.26 |
ENSMUST00000182935.1
ENSMUST00000182506.1 |
Ccnd3
|
cyclin D3 |
| chr6_-_3968357 | 0.26 |
ENSMUST00000031674.8
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr11_+_94211431 | 0.26 |
ENSMUST00000041589.5
|
Tob1
|
transducer of ErbB-2.1 |
| chr16_-_33380717 | 0.26 |
ENSMUST00000180923.1
|
1700007L15Rik
|
RIKEN cDNA 1700007L15 gene |
| chr14_-_62456286 | 0.25 |
ENSMUST00000165651.1
ENSMUST00000022501.3 |
Gucy1b2
|
guanylate cyclase 1, soluble, beta 2 |
| chr6_+_34780836 | 0.25 |
ENSMUST00000115012.1
ENSMUST00000115014.1 ENSMUST00000115009.1 |
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr17_+_13234871 | 0.25 |
ENSMUST00000059824.6
|
Smok2b
|
sperm motility kinase 2B |
| chr5_-_145191511 | 0.25 |
ENSMUST00000161845.1
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
| chr10_+_7832457 | 0.25 |
ENSMUST00000039484.4
|
Zc3h12d
|
zinc finger CCCH type containing 12D |
| chrX_-_70365052 | 0.25 |
ENSMUST00000101509.2
|
Ids
|
iduronate 2-sulfatase |
| chr8_+_45069137 | 0.25 |
ENSMUST00000067984.7
|
Mtnr1a
|
melatonin receptor 1A |
| chr17_-_35838259 | 0.25 |
ENSMUST00000001566.8
|
Tubb5
|
tubulin, beta 5 class I |
| chr9_+_3335470 | 0.24 |
ENSMUST00000053407.5
|
Alkbh8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr12_-_102757765 | 0.23 |
ENSMUST00000179306.1
ENSMUST00000173969.1 ENSMUST00000179263.1 ENSMUST00000046456.6 |
RP24-234J3.4
AK010878
|
Protein AK010878 cDNA sequence AK010878 |
| chr9_-_75409352 | 0.23 |
ENSMUST00000168937.1
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr6_-_57692007 | 0.23 |
ENSMUST00000185014.1
ENSMUST00000053386.5 |
PYURF
Pyurf
|
protein preY, mitochondrial precursor Pigy upstream reading frame |
| chr4_-_129696817 | 0.23 |
ENSMUST00000102588.3
|
Tmem39b
|
transmembrane protein 39b |
| chr15_-_81926148 | 0.23 |
ENSMUST00000023113.5
|
Polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chrM_+_7759 | 0.23 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr14_+_11227511 | 0.22 |
ENSMUST00000080237.3
|
Rpl21-ps4
|
ribosomal protein L21, pseudogene 4 |
| chr12_+_69296676 | 0.22 |
ENSMUST00000021362.4
|
Klhdc2
|
kelch domain containing 2 |
| chr3_-_64509735 | 0.22 |
ENSMUST00000177184.1
|
Vmn2r5
|
vomeronasal 2, receptor 5 |
| chr12_+_52097737 | 0.22 |
ENSMUST00000040090.9
|
Nubpl
|
nucleotide binding protein-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.7 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.2 | 1.4 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 1.6 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 1.4 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 | 0.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.6 | GO:0060295 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.4 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.3 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.6 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.3 | GO:1904446 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.9 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.6 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.0 | 0.1 | GO:0071613 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.7 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 1.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.3 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 2.1 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 1.3 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.6 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0099578 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 2.1 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.1 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.0 | 0.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.0 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 0.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.4 | GO:1990023 | condensed chromosome outer kinetochore(GO:0000940) mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 2.1 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.3 | 1.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.7 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 0.9 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 1.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 0.4 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 0.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.3 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.5 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |