Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
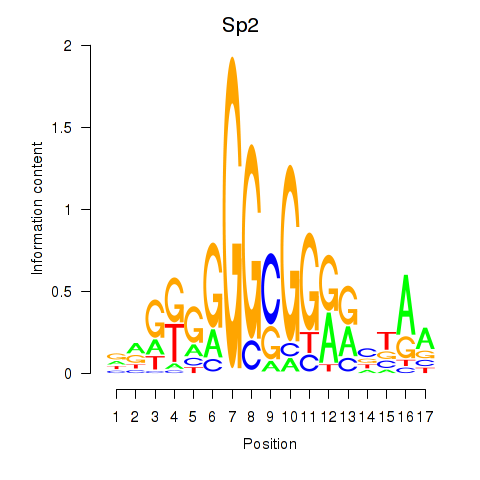
Results for Sp2
Z-value: 0.55
Transcription factors associated with Sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp2
|
ENSMUSG00000018678.6 | Sp2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp2 | mm10_v2_chr11_-_96977660_96977711 | -0.60 | 1.1e-04 | Click! |
Activity profile of Sp2 motif
Sorted Z-values of Sp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_76696725 | 1.53 |
ENSMUST00000023203.4
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr17_-_32917048 | 1.45 |
ENSMUST00000054174.7
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr17_-_32917320 | 1.45 |
ENSMUST00000179434.1
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr15_-_3583146 | 1.06 |
ENSMUST00000110698.2
|
Ghr
|
growth hormone receptor |
| chr10_+_128194446 | 1.06 |
ENSMUST00000044776.6
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr7_-_97579382 | 1.06 |
ENSMUST00000151840.1
ENSMUST00000135998.1 ENSMUST00000144858.1 ENSMUST00000146605.1 ENSMUST00000072725.5 ENSMUST00000138060.1 ENSMUST00000154853.1 ENSMUST00000136757.1 ENSMUST00000124552.1 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr10_+_128194631 | 1.02 |
ENSMUST00000123291.1
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr2_+_119351222 | 0.99 |
ENSMUST00000028780.3
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr5_-_65435717 | 0.87 |
ENSMUST00000117542.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr5_-_65435881 | 0.86 |
ENSMUST00000031103.7
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr10_-_24927444 | 0.82 |
ENSMUST00000020161.8
|
Arg1
|
arginase, liver |
| chr1_-_182409020 | 0.81 |
ENSMUST00000097444.1
|
Gm10517
|
predicted gene 10517 |
| chr9_+_119357381 | 0.79 |
ENSMUST00000039610.8
|
Xylb
|
xylulokinase homolog (H. influenzae) |
| chr1_+_72824482 | 0.79 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr14_+_21052574 | 0.78 |
ENSMUST00000045376.9
|
Adk
|
adenosine kinase |
| chr2_+_31470207 | 0.77 |
ENSMUST00000102840.4
|
Ass1
|
argininosuccinate synthetase 1 |
| chr12_+_78226627 | 0.76 |
ENSMUST00000110388.2
ENSMUST00000052472.4 |
Gphn
|
gephyrin |
| chr1_+_131962941 | 0.70 |
ENSMUST00000177943.1
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr11_+_16752203 | 0.70 |
ENSMUST00000102884.3
ENSMUST00000020329.6 |
Egfr
|
epidermal growth factor receptor |
| chr9_+_121642716 | 0.69 |
ENSMUST00000035115.4
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr16_+_20733104 | 0.68 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr5_+_134986191 | 0.67 |
ENSMUST00000094245.2
|
Cldn3
|
claudin 3 |
| chr6_+_125321409 | 0.66 |
ENSMUST00000176442.1
ENSMUST00000177329.1 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr14_-_31640878 | 0.60 |
ENSMUST00000167066.1
ENSMUST00000127204.2 ENSMUST00000022437.8 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr11_+_94211431 | 0.60 |
ENSMUST00000041589.5
|
Tob1
|
transducer of ErbB-2.1 |
| chr7_-_45061706 | 0.59 |
ENSMUST00000107832.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr17_-_45686120 | 0.59 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr3_-_101604580 | 0.58 |
ENSMUST00000036493.6
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr13_-_29984219 | 0.56 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr17_+_24804312 | 0.52 |
ENSMUST00000024972.5
|
Meiob
|
meiosis specific with OB domains |
| chr7_-_43489967 | 0.51 |
ENSMUST00000107974.1
|
Iglon5
|
IgLON family member 5 |
| chr7_-_45061651 | 0.49 |
ENSMUST00000007981.3
ENSMUST00000107831.1 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr11_+_98348404 | 0.48 |
ENSMUST00000078694.6
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr11_-_74925925 | 0.47 |
ENSMUST00000121738.1
|
Srr
|
serine racemase |
| chr1_-_65179058 | 0.47 |
ENSMUST00000097709.4
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr12_+_8771317 | 0.46 |
ENSMUST00000020911.7
|
Sdc1
|
syndecan 1 |
| chr15_+_55112420 | 0.46 |
ENSMUST00000100660.4
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr18_-_16809233 | 0.46 |
ENSMUST00000025166.7
|
Cdh2
|
cadherin 2 |
| chr12_+_41024090 | 0.45 |
ENSMUST00000132121.1
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_+_55112317 | 0.45 |
ENSMUST00000096433.3
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr12_-_35534973 | 0.45 |
ENSMUST00000116436.2
|
Ahr
|
aryl-hydrocarbon receptor |
| chr6_-_120294559 | 0.45 |
ENSMUST00000057283.7
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr12_+_41024329 | 0.45 |
ENSMUST00000134965.1
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr11_+_119942763 | 0.44 |
ENSMUST00000026436.3
ENSMUST00000106231.1 ENSMUST00000075180.5 ENSMUST00000103021.3 ENSMUST00000106233.1 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr16_-_10543028 | 0.44 |
ENSMUST00000184863.1
ENSMUST00000038281.5 |
Dexi
|
dexamethasone-induced transcript |
| chr1_-_184033998 | 0.43 |
ENSMUST00000050306.5
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr1_-_191397026 | 0.43 |
ENSMUST00000067976.3
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B (B56), alpha isoform |
| chr7_+_30231884 | 0.42 |
ENSMUST00000019882.9
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr1_-_65186456 | 0.41 |
ENSMUST00000169032.1
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr1_+_184034381 | 0.41 |
ENSMUST00000048655.7
|
Dusp10
|
dual specificity phosphatase 10 |
| chr14_-_61556746 | 0.40 |
ENSMUST00000100496.4
|
Spryd7
|
SPRY domain containing 7 |
| chr11_+_79660532 | 0.40 |
ENSMUST00000155381.1
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chrX_-_169320273 | 0.40 |
ENSMUST00000033717.2
ENSMUST00000112115.1 |
Hccs
|
holocytochrome c synthetase |
| chr4_+_43046014 | 0.40 |
ENSMUST00000180426.1
|
Gm26881
|
predicted gene, 26881 |
| chrX_+_142228177 | 0.40 |
ENSMUST00000112914.1
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr10_-_59616667 | 0.39 |
ENSMUST00000020312.6
|
Mcu
|
mitochondrial calcium uniporter |
| chr15_-_58214882 | 0.39 |
ENSMUST00000022986.6
|
Fbxo32
|
F-box protein 32 |
| chr5_+_72914554 | 0.39 |
ENSMUST00000143829.1
|
Slain2
|
SLAIN motif family, member 2 |
| chr17_-_45686214 | 0.38 |
ENSMUST00000113523.2
|
Tmem63b
|
transmembrane protein 63b |
| chr14_+_31641051 | 0.38 |
ENSMUST00000090147.6
|
Btd
|
biotinidase |
| chr4_+_41760454 | 0.38 |
ENSMUST00000108040.1
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr11_+_78194696 | 0.38 |
ENSMUST00000060539.6
|
Proca1
|
protein interacting with cyclin A1 |
| chr4_+_116720920 | 0.37 |
ENSMUST00000045542.6
ENSMUST00000106459.1 |
Tesk2
|
testis-specific kinase 2 |
| chr1_-_130715734 | 0.37 |
ENSMUST00000066863.6
ENSMUST00000050406.4 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr11_-_21572193 | 0.37 |
ENSMUST00000102874.4
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr10_+_29143996 | 0.36 |
ENSMUST00000092629.2
|
Soga3
|
SOGA family member 3 |
| chr19_-_45006385 | 0.36 |
ENSMUST00000097715.2
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr5_+_30814571 | 0.36 |
ENSMUST00000031058.8
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr6_-_119848059 | 0.36 |
ENSMUST00000184864.1
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr11_-_101171302 | 0.36 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_-_45595842 | 0.36 |
ENSMUST00000164618.1
ENSMUST00000097317.3 ENSMUST00000170113.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr11_-_115187321 | 0.36 |
ENSMUST00000103038.1
ENSMUST00000103039.1 ENSMUST00000103040.4 |
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_+_30364262 | 0.36 |
ENSMUST00000142801.1
ENSMUST00000100214.3 |
Fam73b
|
family with sequence similarity 73, member B |
| chr15_-_54920115 | 0.35 |
ENSMUST00000171545.1
|
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_+_30814722 | 0.35 |
ENSMUST00000114724.1
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr15_-_54919961 | 0.35 |
ENSMUST00000167541.2
ENSMUST00000041591.9 ENSMUST00000173516.1 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr16_-_20621255 | 0.35 |
ENSMUST00000052939.2
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chrX_+_142227923 | 0.34 |
ENSMUST00000042329.5
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_30364227 | 0.34 |
ENSMUST00000077977.7
ENSMUST00000140075.2 |
Fam73b
|
family with sequence similarity 73, member B |
| chr14_-_61556881 | 0.34 |
ENSMUST00000022497.8
|
Spryd7
|
SPRY domain containing 7 |
| chr11_-_102296618 | 0.34 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr2_-_84775388 | 0.34 |
ENSMUST00000023994.3
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr19_+_6497772 | 0.34 |
ENSMUST00000113458.1
ENSMUST00000113459.1 |
Nrxn2
|
neurexin II |
| chr8_-_61591130 | 0.34 |
ENSMUST00000135439.1
ENSMUST00000121200.1 |
Palld
|
palladin, cytoskeletal associated protein |
| chr17_+_45686322 | 0.33 |
ENSMUST00000024734.7
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr16_+_45158725 | 0.33 |
ENSMUST00000023343.3
|
Atg3
|
autophagy related 3 |
| chr11_+_101468164 | 0.33 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr14_+_118937925 | 0.33 |
ENSMUST00000022734.7
|
Dnajc3
|
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr14_-_21052452 | 0.33 |
ENSMUST00000130291.1
|
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr9_-_36797273 | 0.33 |
ENSMUST00000163192.3
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr11_-_115187827 | 0.33 |
ENSMUST00000103041.1
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr6_-_119848093 | 0.32 |
ENSMUST00000079582.4
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_+_147187424 | 0.32 |
ENSMUST00000144411.1
|
6430503K07Rik
|
RIKEN cDNA 6430503K07 gene |
| chr17_-_45685973 | 0.32 |
ENSMUST00000145873.1
|
Tmem63b
|
transmembrane protein 63b |
| chr4_-_114908892 | 0.32 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr6_-_119848120 | 0.32 |
ENSMUST00000183703.1
ENSMUST00000183911.1 |
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr19_+_42090422 | 0.31 |
ENSMUST00000066778.4
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr9_-_31211805 | 0.31 |
ENSMUST00000072634.7
ENSMUST00000079758.7 |
Aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr3_+_29082539 | 0.31 |
ENSMUST00000119598.1
ENSMUST00000118531.1 |
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr18_+_64340225 | 0.31 |
ENSMUST00000175965.2
ENSMUST00000115145.3 |
Onecut2
|
one cut domain, family member 2 |
| chr7_-_25658726 | 0.31 |
ENSMUST00000071329.6
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr12_-_108275409 | 0.31 |
ENSMUST00000136175.1
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr11_+_78194734 | 0.30 |
ENSMUST00000108317.2
|
Proca1
|
protein interacting with cyclin A1 |
| chr6_+_37870786 | 0.30 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr2_-_84775420 | 0.30 |
ENSMUST00000111641.1
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr2_-_161109017 | 0.30 |
ENSMUST00000039782.7
ENSMUST00000134178.1 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr9_+_59578192 | 0.30 |
ENSMUST00000118549.1
ENSMUST00000034840.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chrX_+_161717498 | 0.30 |
ENSMUST00000061514.7
|
Rai2
|
retinoic acid induced 2 |
| chrX_-_48208566 | 0.30 |
ENSMUST00000037960.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr3_+_90080442 | 0.30 |
ENSMUST00000127955.1
|
Tpm3
|
tropomyosin 3, gamma |
| chr1_-_179546261 | 0.30 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr4_-_125065603 | 0.29 |
ENSMUST00000036383.3
|
Dnali1
|
dynein, axonemal, light intermediate polypeptide 1 |
| chr14_+_118137101 | 0.29 |
ENSMUST00000022728.2
|
Gpr180
|
G protein-coupled receptor 180 |
| chr11_+_84179852 | 0.29 |
ENSMUST00000136463.2
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr8_-_104395765 | 0.29 |
ENSMUST00000179802.1
|
Cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr14_-_21052120 | 0.29 |
ENSMUST00000130370.1
ENSMUST00000022371.3 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr1_-_162937225 | 0.29 |
ENSMUST00000178465.1
|
Fmo6
|
flavin containing monooxygenase 6 |
| chr7_-_63938862 | 0.29 |
ENSMUST00000063694.8
|
Klf13
|
Kruppel-like factor 13 |
| chr10_+_13966268 | 0.28 |
ENSMUST00000015645.4
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chrX_+_161717055 | 0.28 |
ENSMUST00000112338.1
|
Rai2
|
retinoic acid induced 2 |
| chr8_+_94037198 | 0.28 |
ENSMUST00000109556.2
ENSMUST00000093301.2 ENSMUST00000060632.7 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr5_+_104459450 | 0.28 |
ENSMUST00000086831.3
|
Pkd2
|
polycystic kidney disease 2 |
| chr4_+_117251951 | 0.28 |
ENSMUST00000062824.5
|
Tmem53
|
transmembrane protein 53 |
| chr11_-_97187872 | 0.28 |
ENSMUST00000001479.4
|
Kpnb1
|
karyopherin (importin) beta 1 |
| chr18_+_38993126 | 0.28 |
ENSMUST00000097593.2
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr7_-_34655500 | 0.27 |
ENSMUST00000032709.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr4_+_117252010 | 0.27 |
ENSMUST00000125943.1
ENSMUST00000106434.1 |
Tmem53
|
transmembrane protein 53 |
| chrX_-_56598069 | 0.27 |
ENSMUST00000059899.2
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr16_-_45158624 | 0.27 |
ENSMUST00000180636.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr10_+_127165118 | 0.26 |
ENSMUST00000006914.9
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr4_+_107178399 | 0.26 |
ENSMUST00000030361.4
ENSMUST00000128123.1 ENSMUST00000106753.1 |
Tmem59
|
transmembrane protein 59 |
| chr8_-_106136792 | 0.26 |
ENSMUST00000146940.1
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr11_-_70646972 | 0.26 |
ENSMUST00000014750.8
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr11_-_73177002 | 0.26 |
ENSMUST00000108480.1
ENSMUST00000054952.3 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr9_-_36797303 | 0.26 |
ENSMUST00000115086.5
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr3_-_97767916 | 0.26 |
ENSMUST00000045243.8
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr1_+_151344472 | 0.26 |
ENSMUST00000023918.6
ENSMUST00000097543.1 ENSMUST00000111887.3 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr7_+_44849949 | 0.26 |
ENSMUST00000141311.1
ENSMUST00000107880.1 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr17_-_23786046 | 0.26 |
ENSMUST00000024704.3
|
Flywch2
|
FLYWCH family member 2 |
| chr16_-_45158453 | 0.26 |
ENSMUST00000181750.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr1_+_128244122 | 0.26 |
ENSMUST00000027592.3
|
Ubxn4
|
UBX domain protein 4 |
| chr1_+_66700831 | 0.25 |
ENSMUST00000027157.3
ENSMUST00000113995.1 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr11_-_4160286 | 0.25 |
ENSMUST00000093381.4
ENSMUST00000101626.2 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr1_+_59482133 | 0.25 |
ENSMUST00000114246.2
ENSMUST00000037105.6 |
Fzd7
|
frizzled homolog 7 (Drosophila) |
| chr14_+_30715599 | 0.25 |
ENSMUST00000054230.4
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr18_-_20896078 | 0.25 |
ENSMUST00000025177.6
ENSMUST00000097658.1 |
Trappc8
|
trafficking protein particle complex 8 |
| chr10_-_123076367 | 0.25 |
ENSMUST00000073792.3
ENSMUST00000170935.1 ENSMUST00000037557.7 |
Mon2
|
MON2 homolog (yeast) |
| chr9_+_47530173 | 0.25 |
ENSMUST00000114548.1
ENSMUST00000152459.1 ENSMUST00000143026.1 ENSMUST00000085909.2 ENSMUST00000114547.1 ENSMUST00000034581.3 |
Cadm1
|
cell adhesion molecule 1 |
| chr9_+_83925118 | 0.25 |
ENSMUST00000034801.4
|
Bckdhb
|
branched chain ketoacid dehydrogenase E1, beta polypeptide |
| chr18_-_66022580 | 0.25 |
ENSMUST00000143990.1
|
Lman1
|
lectin, mannose-binding, 1 |
| chr10_-_75798576 | 0.25 |
ENSMUST00000001713.3
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr14_+_73173825 | 0.25 |
ENSMUST00000166875.1
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr4_-_72200833 | 0.24 |
ENSMUST00000102848.2
ENSMUST00000072695.6 ENSMUST00000107337.1 ENSMUST00000074216.7 |
Tle1
|
transducin-like enhancer of split 1, homolog of Drosophila E(spl) |
| chr9_-_106438207 | 0.24 |
ENSMUST00000024031.6
|
Acy1
|
aminoacylase 1 |
| chr7_+_131371138 | 0.24 |
ENSMUST00000075610.6
|
Pstk
|
phosphoseryl-tRNA kinase |
| chr14_-_122983142 | 0.24 |
ENSMUST00000126867.1
ENSMUST00000148661.1 |
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chrX_-_36645359 | 0.24 |
ENSMUST00000051906.6
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr1_+_106171752 | 0.24 |
ENSMUST00000061047.6
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr15_+_99295087 | 0.23 |
ENSMUST00000128352.1
ENSMUST00000145482.1 |
Prpf40b
|
PRP40 pre-mRNA processing factor 40 homolog B (yeast) |
| chr7_+_30232032 | 0.23 |
ENSMUST00000149654.1
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr7_-_74554474 | 0.23 |
ENSMUST00000134539.1
ENSMUST00000026897.7 ENSMUST00000098371.2 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr1_-_93478785 | 0.23 |
ENSMUST00000170883.1
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr7_-_89338709 | 0.23 |
ENSMUST00000137723.1
ENSMUST00000117852.1 ENSMUST00000041968.3 |
Tmem135
|
transmembrane protein 135 |
| chr9_-_103365769 | 0.23 |
ENSMUST00000035484.4
ENSMUST00000072249.6 |
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr3_-_88425094 | 0.23 |
ENSMUST00000168755.1
ENSMUST00000057935.6 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr12_-_91849081 | 0.23 |
ENSMUST00000167466.1
ENSMUST00000021347.5 ENSMUST00000178462.1 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr14_+_28504736 | 0.23 |
ENSMUST00000063465.4
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr5_+_144255223 | 0.23 |
ENSMUST00000056578.6
|
Bri3
|
brain protein I3 |
| chr5_-_140830430 | 0.23 |
ENSMUST00000000153.4
|
Gna12
|
guanine nucleotide binding protein, alpha 12 |
| chr14_-_122983211 | 0.23 |
ENSMUST00000037726.7
|
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr9_+_21955747 | 0.22 |
ENSMUST00000053583.5
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr7_+_96951552 | 0.22 |
ENSMUST00000107159.1
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr16_-_45158650 | 0.22 |
ENSMUST00000023344.3
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr17_+_85621017 | 0.22 |
ENSMUST00000162695.2
|
Six3
|
sine oculis-related homeobox 3 |
| chr4_-_130275523 | 0.22 |
ENSMUST00000146478.1
|
Serinc2
|
serine incorporator 2 |
| chr7_-_27166732 | 0.22 |
ENSMUST00000080058.4
|
Egln2
|
EGL nine homolog 2 (C. elegans) |
| chr14_+_21500879 | 0.22 |
ENSMUST00000182964.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr5_-_147076482 | 0.22 |
ENSMUST00000016664.7
|
Lnx2
|
ligand of numb-protein X 2 |
| chr12_-_31950170 | 0.22 |
ENSMUST00000176520.1
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr16_-_45158566 | 0.22 |
ENSMUST00000181177.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr13_+_81711407 | 0.22 |
ENSMUST00000057598.5
|
Mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr6_+_124931378 | 0.22 |
ENSMUST00000032214.7
ENSMUST00000180095.1 |
Mlf2
|
myeloid leukemia factor 2 |
| chrX_-_38564519 | 0.22 |
ENSMUST00000016681.8
|
Cul4b
|
cullin 4B |
| chr7_-_101933815 | 0.22 |
ENSMUST00000106963.1
ENSMUST00000106966.1 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr5_+_129941949 | 0.21 |
ENSMUST00000051758.7
ENSMUST00000073945.4 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr4_-_151108454 | 0.21 |
ENSMUST00000105670.1
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr13_-_34130345 | 0.21 |
ENSMUST00000075774.3
|
Tubb2b
|
tubulin, beta 2B class IIB |
| chr4_-_130275542 | 0.21 |
ENSMUST00000154846.1
ENSMUST00000105996.1 |
Serinc2
|
serine incorporator 2 |
| chr11_+_114851507 | 0.21 |
ENSMUST00000177952.1
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr4_+_62360695 | 0.21 |
ENSMUST00000084526.5
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr5_-_123749393 | 0.21 |
ENSMUST00000057795.5
ENSMUST00000111515.1 ENSMUST00000182309.1 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr7_-_101933780 | 0.21 |
ENSMUST00000106964.1
ENSMUST00000078448.3 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr4_-_117133953 | 0.21 |
ENSMUST00000076859.5
|
Plk3
|
polo-like kinase 3 |
| chr14_-_55591077 | 0.21 |
ENSMUST00000161807.1
ENSMUST00000111378.3 ENSMUST00000159687.1 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr11_-_116138862 | 0.21 |
ENSMUST00000106439.1
|
Mrpl38
|
mitochondrial ribosomal protein L38 |
| chr2_+_19658055 | 0.21 |
ENSMUST00000052168.4
|
Otud1
|
OTU domain containing 1 |
| chr7_+_96951505 | 0.21 |
ENSMUST00000044466.5
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr7_-_45717890 | 0.21 |
ENSMUST00000107738.3
|
Sphk2
|
sphingosine kinase 2 |
| chrX_+_94234594 | 0.21 |
ENSMUST00000153900.1
|
Klhl15
|
kelch-like 15 |
| chrX_+_94367147 | 0.21 |
ENSMUST00000113897.2
ENSMUST00000113896.1 ENSMUST00000113895.1 |
Apoo
|
apolipoprotein O |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.3 | 0.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 0.9 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 0.8 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.3 | 0.8 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.2 | 0.7 | GO:0001192 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.2 | 0.6 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 0.8 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.8 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.2 | 0.7 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 0.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.5 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 0.9 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.6 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.4 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 0.3 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.1 | 0.4 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.3 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 1.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.1 | 0.3 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.5 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.1 | 0.3 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.3 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.3 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.2 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.3 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.7 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:0061114 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.2 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.1 | 1.0 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.2 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 | 1.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.2 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.1 | GO:0051586 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.6 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.2 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0007228 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 2.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 1.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.4 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) regulation of cardiac ventricle development(GO:1904412) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.0 | 0.1 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.4 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:1904714 | Golgi disassembly(GO:0090166) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.7 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.4 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.2 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 1.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.0 | 0.0 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.2 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 0.9 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.3 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 2.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.6 | 1.7 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.4 | 1.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 2.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 0.9 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 0.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.2 | 0.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 1.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.7 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.7 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.5 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.2 | 0.5 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.7 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.6 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.4 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.1 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.1 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 1.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.6 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |