Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
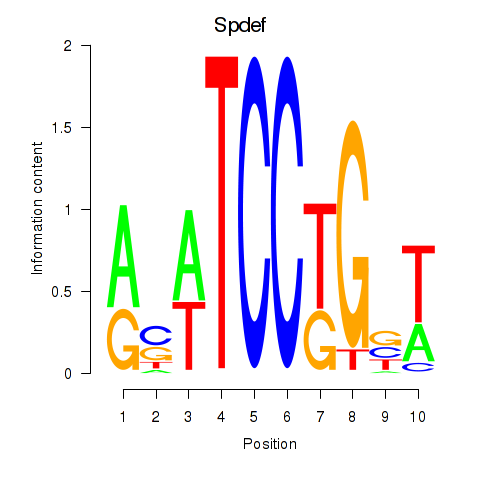
Results for Spdef
Z-value: 0.98
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSMUSG00000024215.7 | SAM pointed domain containing ets transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spdef | mm10_v2_chr17_-_27728889_27728956 | -0.16 | 3.6e-01 | Click! |
Activity profile of Spdef motif
Sorted Z-values of Spdef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_40025253 | 3.66 |
ENSMUST00000163705.2
|
AI317395
|
expressed sequence AI317395 |
| chr8_+_105269837 | 3.58 |
ENSMUST00000172525.1
ENSMUST00000174837.1 ENSMUST00000173859.1 |
Hsf4
|
heat shock transcription factor 4 |
| chr8_+_105269788 | 3.50 |
ENSMUST00000036127.2
ENSMUST00000163734.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr7_+_87246649 | 2.75 |
ENSMUST00000068829.5
ENSMUST00000032781.7 |
Nox4
|
NADPH oxidase 4 |
| chr9_-_65908676 | 2.15 |
ENSMUST00000119245.1
ENSMUST00000134338.1 ENSMUST00000179395.1 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr15_-_89170688 | 2.10 |
ENSMUST00000060808.9
|
Plxnb2
|
plexin B2 |
| chrX_-_75843063 | 2.01 |
ENSMUST00000114057.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr1_-_121328024 | 1.97 |
ENSMUST00000003818.7
|
Insig2
|
insulin induced gene 2 |
| chrX_-_75843185 | 1.92 |
ENSMUST00000137192.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr7_+_143473736 | 1.78 |
ENSMUST00000052348.5
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr9_+_108296853 | 1.66 |
ENSMUST00000035230.5
|
Amt
|
aminomethyltransferase |
| chr18_+_12643329 | 1.65 |
ENSMUST00000025294.7
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr17_-_32947372 | 1.62 |
ENSMUST00000139353.1
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr5_-_25100624 | 1.56 |
ENSMUST00000030784.7
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_-_105944412 | 1.54 |
ENSMUST00000019734.4
ENSMUST00000184269.1 ENSMUST00000150563.1 |
Cyb561
|
cytochrome b-561 |
| chr19_+_41029275 | 1.50 |
ENSMUST00000051806.4
ENSMUST00000112200.1 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr19_-_10830045 | 1.50 |
ENSMUST00000080292.5
|
Cd6
|
CD6 antigen |
| chr10_-_95324072 | 1.50 |
ENSMUST00000053594.5
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_-_89393629 | 1.42 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr15_+_102102926 | 1.38 |
ENSMUST00000169627.1
ENSMUST00000046144.9 |
Tenc1
|
tensin like C1 domain-containing phosphatase |
| chr18_-_3281036 | 1.38 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr9_-_106476590 | 1.31 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr16_+_5007283 | 1.30 |
ENSMUST00000184439.1
|
Smim22
|
small integral membrane protein 22 |
| chr19_-_10829921 | 1.29 |
ENSMUST00000039043.8
|
Cd6
|
CD6 antigen |
| chr13_-_41847482 | 1.28 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr1_+_135818593 | 1.28 |
ENSMUST00000038760.8
|
Lad1
|
ladinin |
| chr19_-_29047847 | 1.28 |
ENSMUST00000025696.4
|
Ak3
|
adenylate kinase 3 |
| chr17_+_32506446 | 1.25 |
ENSMUST00000165999.1
|
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr9_-_50746501 | 1.23 |
ENSMUST00000034564.1
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr13_+_40886758 | 1.23 |
ENSMUST00000069958.7
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr16_-_44016387 | 1.21 |
ENSMUST00000036174.3
|
Gramd1c
|
GRAM domain containing 1C |
| chr2_+_72054598 | 1.21 |
ENSMUST00000028525.5
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_45705088 | 1.19 |
ENSMUST00000080885.3
|
Dbp
|
D site albumin promoter binding protein |
| chr3_-_82903963 | 1.19 |
ENSMUST00000029632.6
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr3_+_135825788 | 1.18 |
ENSMUST00000167390.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr4_-_107923519 | 1.14 |
ENSMUST00000106719.1
ENSMUST00000106720.2 ENSMUST00000131644.1 ENSMUST00000030345.8 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr7_+_101378183 | 1.13 |
ENSMUST00000084895.5
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_-_20112876 | 1.12 |
ENSMUST00000000137.7
|
Actr2
|
ARP2 actin-related protein 2 |
| chrX_+_107255878 | 1.11 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr4_-_130279205 | 1.09 |
ENSMUST00000120126.2
|
Serinc2
|
serine incorporator 2 |
| chr11_-_72795801 | 1.08 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr19_-_10829856 | 1.05 |
ENSMUST00000174176.1
|
Cd6
|
CD6 antigen |
| chr12_-_28623282 | 1.05 |
ENSMUST00000036136.7
|
Colec11
|
collectin sub-family member 11 |
| chr2_-_163419508 | 1.02 |
ENSMUST00000046908.3
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr5_+_129941949 | 1.00 |
ENSMUST00000051758.7
ENSMUST00000073945.4 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr2_-_73386396 | 0.98 |
ENSMUST00000112044.1
ENSMUST00000112043.1 ENSMUST00000076463.5 |
Gpr155
|
G protein-coupled receptor 155 |
| chr10_+_34483400 | 0.98 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr3_-_63964659 | 0.98 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr4_+_155582476 | 0.97 |
ENSMUST00000105612.1
|
Nadk
|
NAD kinase |
| chr11_-_70646972 | 0.97 |
ENSMUST00000014750.8
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr6_-_95718800 | 0.96 |
ENSMUST00000079847.5
|
Suclg2
|
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| chr14_+_53743104 | 0.94 |
ENSMUST00000103667.4
|
Trav16
|
T cell receptor alpha variable 16 |
| chr3_-_5576111 | 0.94 |
ENSMUST00000165309.1
ENSMUST00000164828.1 ENSMUST00000071280.5 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr8_+_72161101 | 0.94 |
ENSMUST00000003121.8
|
Rab8a
|
RAB8A, member RAS oncogene family |
| chr6_-_87690819 | 0.94 |
ENSMUST00000162547.1
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene |
| chr15_+_88819584 | 0.94 |
ENSMUST00000024042.3
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr4_+_140961203 | 0.93 |
ENSMUST00000010007.8
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr6_-_57692007 | 0.92 |
ENSMUST00000185014.1
ENSMUST00000053386.5 |
PYURF
Pyurf
|
protein preY, mitochondrial precursor Pigy upstream reading frame |
| chr4_+_80910646 | 0.91 |
ENSMUST00000055922.3
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr1_+_84839833 | 0.91 |
ENSMUST00000097672.3
|
Fbxo36
|
F-box protein 36 |
| chr13_+_40859768 | 0.91 |
ENSMUST00000110191.2
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr15_+_58933774 | 0.91 |
ENSMUST00000022980.3
|
Ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chrX_+_42502533 | 0.90 |
ENSMUST00000005839.4
|
Sh2d1a
|
SH2 domain protein 1A |
| chr8_-_64849818 | 0.90 |
ENSMUST00000034017.7
|
Klhl2
|
kelch-like 2, Mayven |
| chr15_+_94543605 | 0.89 |
ENSMUST00000074936.3
|
Irak4
|
interleukin-1 receptor-associated kinase 4 |
| chr17_-_57247632 | 0.89 |
ENSMUST00000005975.6
|
Gpr108
|
G protein-coupled receptor 108 |
| chr11_-_69685537 | 0.89 |
ENSMUST00000018896.7
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_89393294 | 0.88 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr3_-_5576233 | 0.88 |
ENSMUST00000059021.4
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr19_+_8920358 | 0.88 |
ENSMUST00000096243.5
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr2_+_152669461 | 0.87 |
ENSMUST00000125366.1
ENSMUST00000109825.1 ENSMUST00000089059.2 ENSMUST00000079247.3 |
H13
|
histocompatibility 13 |
| chr11_+_87592145 | 0.87 |
ENSMUST00000103179.3
ENSMUST00000092802.5 ENSMUST00000146871.1 |
Mtmr4
|
myotubularin related protein 4 |
| chr9_+_80165013 | 0.87 |
ENSMUST00000035889.8
ENSMUST00000113268.1 |
Myo6
|
myosin VI |
| chr11_-_21572193 | 0.87 |
ENSMUST00000102874.4
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr6_-_119388671 | 0.85 |
ENSMUST00000169744.1
|
Adipor2
|
adiponectin receptor 2 |
| chr12_+_76533540 | 0.85 |
ENSMUST00000075249.4
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_+_95893953 | 0.84 |
ENSMUST00000056710.8
ENSMUST00000015894.5 |
Aph1a
|
anterior pharynx defective 1a homolog (C. elegans) |
| chr11_-_53430417 | 0.84 |
ENSMUST00000109019.1
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr3_+_87919563 | 0.83 |
ENSMUST00000121920.1
|
Mrpl24
|
mitochondrial ribosomal protein L24 |
| chr5_-_140830430 | 0.81 |
ENSMUST00000000153.4
|
Gna12
|
guanine nucleotide binding protein, alpha 12 |
| chr7_+_19965365 | 0.81 |
ENSMUST00000094753.4
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr9_+_6168638 | 0.80 |
ENSMUST00000058692.7
|
Pdgfd
|
platelet-derived growth factor, D polypeptide |
| chr4_+_43562672 | 0.80 |
ENSMUST00000167751.1
ENSMUST00000132631.1 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr11_+_70092653 | 0.80 |
ENSMUST00000143772.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr2_+_145785980 | 0.80 |
ENSMUST00000110005.1
ENSMUST00000094480.4 |
Rin2
|
Ras and Rab interactor 2 |
| chr15_-_37458523 | 0.79 |
ENSMUST00000116445.2
|
Ncald
|
neurocalcin delta |
| chr11_+_70092634 | 0.79 |
ENSMUST00000102572.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr8_-_124569696 | 0.79 |
ENSMUST00000063278.6
|
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr1_-_183297008 | 0.78 |
ENSMUST00000057062.5
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr6_+_29468068 | 0.78 |
ENSMUST00000143101.1
|
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr16_-_87432597 | 0.78 |
ENSMUST00000039449.7
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr2_-_160872985 | 0.78 |
ENSMUST00000109460.1
ENSMUST00000127201.1 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr13_-_60177357 | 0.77 |
ENSMUST00000065086.4
|
Gas1
|
growth arrest specific 1 |
| chr9_+_110476985 | 0.77 |
ENSMUST00000084948.4
ENSMUST00000061155.6 ENSMUST00000140686.1 ENSMUST00000084952.5 |
Kif9
|
kinesin family member 9 |
| chr19_+_6363719 | 0.77 |
ENSMUST00000113489.1
ENSMUST00000113488.1 |
Sf1
|
splicing factor 1 |
| chr8_+_22283441 | 0.76 |
ENSMUST00000077194.1
|
Tpte
|
transmembrane phosphatase with tensin homology |
| chr2_-_18392736 | 0.76 |
ENSMUST00000091418.5
ENSMUST00000166495.1 |
Dnajc1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr9_+_80165079 | 0.76 |
ENSMUST00000184480.1
|
Myo6
|
myosin VI |
| chr9_+_119402444 | 0.75 |
ENSMUST00000035093.8
ENSMUST00000165044.1 |
Acvr2b
|
activin receptor IIB |
| chr19_+_6363671 | 0.75 |
ENSMUST00000131252.1
|
Sf1
|
splicing factor 1 |
| chr5_+_105732063 | 0.75 |
ENSMUST00000154807.1
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr16_+_5007306 | 0.75 |
ENSMUST00000178155.2
ENSMUST00000184256.1 ENSMUST00000185147.1 |
Smim22
|
small integral membrane protein 22 |
| chr3_+_87919490 | 0.75 |
ENSMUST00000019854.6
ENSMUST00000119968.1 |
Mrpl24
|
mitochondrial ribosomal protein L24 |
| chr11_+_70092705 | 0.74 |
ENSMUST00000124721.1
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chrX_-_134276888 | 0.72 |
ENSMUST00000113252.1
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr11_-_5542177 | 0.72 |
ENSMUST00000020776.4
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr2_+_137663424 | 0.72 |
ENSMUST00000134833.1
|
Gm14064
|
predicted gene 14064 |
| chr1_+_138963709 | 0.71 |
ENSMUST00000168527.1
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr2_-_12419456 | 0.71 |
ENSMUST00000154899.1
ENSMUST00000028105.6 |
Fam188a
|
family with sequence similarity 188, member A |
| chr3_+_90051630 | 0.71 |
ENSMUST00000159064.1
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chrX_+_161717498 | 0.70 |
ENSMUST00000061514.7
|
Rai2
|
retinoic acid induced 2 |
| chr5_+_105731755 | 0.69 |
ENSMUST00000127686.1
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr17_+_34670535 | 0.69 |
ENSMUST00000168533.1
ENSMUST00000087399.4 |
Tnxb
|
tenascin XB |
| chr19_+_6047081 | 0.68 |
ENSMUST00000025723.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr3_-_63964768 | 0.68 |
ENSMUST00000029402.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr7_-_80232479 | 0.67 |
ENSMUST00000123279.1
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr9_+_121898458 | 0.67 |
ENSMUST00000050327.3
|
Ackr2
|
atypical chemokine receptor 2 |
| chr14_+_28511344 | 0.66 |
ENSMUST00000112272.1
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr7_-_73537621 | 0.65 |
ENSMUST00000172704.1
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr8_+_72219726 | 0.65 |
ENSMUST00000003123.8
|
Fam32a
|
family with sequence similarity 32, member A |
| chr3_-_95739544 | 0.64 |
ENSMUST00000153026.1
ENSMUST00000123143.1 ENSMUST00000137912.1 ENSMUST00000029753.6 ENSMUST00000131376.1 ENSMUST00000117507.2 ENSMUST00000128885.1 ENSMUST00000147217.1 |
Ecm1
|
extracellular matrix protein 1 |
| chr9_+_6168601 | 0.64 |
ENSMUST00000168039.1
|
Pdgfd
|
platelet-derived growth factor, D polypeptide |
| chr6_+_29467718 | 0.64 |
ENSMUST00000004396.6
|
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr19_+_6047055 | 0.64 |
ENSMUST00000134667.1
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr11_+_105975204 | 0.63 |
ENSMUST00000001964.7
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr14_-_70207637 | 0.63 |
ENSMUST00000022682.5
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr3_-_84582476 | 0.63 |
ENSMUST00000107687.2
ENSMUST00000098990.3 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr11_+_117232254 | 0.63 |
ENSMUST00000106354.2
|
Sept9
|
septin 9 |
| chr2_-_160872829 | 0.62 |
ENSMUST00000176141.1
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr19_+_6046576 | 0.62 |
ENSMUST00000138532.1
ENSMUST00000129081.1 ENSMUST00000156550.1 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr10_-_95416850 | 0.61 |
ENSMUST00000020215.9
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr8_+_105305572 | 0.61 |
ENSMUST00000109375.2
|
Elmo3
|
engulfment and cell motility 3 |
| chrX_-_134276969 | 0.61 |
ENSMUST00000087541.5
ENSMUST00000087540.3 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr9_+_30427329 | 0.61 |
ENSMUST00000164099.1
|
Snx19
|
sorting nexin 19 |
| chr9_+_123529843 | 0.61 |
ENSMUST00000026270.7
|
Sacm1l
|
SAC1 (suppressor of actin mutations 1, homolog)-like (S. cerevisiae) |
| chr2_+_6322621 | 0.61 |
ENSMUST00000114937.1
|
Usp6nl
|
USP6 N-terminal like |
| chr15_-_54278420 | 0.60 |
ENSMUST00000079772.3
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr12_+_80644212 | 0.60 |
ENSMUST00000085245.5
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr7_-_68353182 | 0.60 |
ENSMUST00000123509.1
ENSMUST00000129965.1 |
Gm16158
|
predicted gene 16158 |
| chr6_+_21986887 | 0.59 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr10_-_128821576 | 0.59 |
ENSMUST00000026409.3
|
Ormdl2
|
ORM1-like 2 (S. cerevisiae) |
| chr14_-_55671873 | 0.59 |
ENSMUST00000163750.1
ENSMUST00000010520.8 |
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr8_+_110847015 | 0.58 |
ENSMUST00000172668.1
ENSMUST00000034203.10 ENSMUST00000174398.1 |
Cog4
|
component of oligomeric golgi complex 4 |
| chr2_+_147012996 | 0.58 |
ENSMUST00000028921.5
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr7_+_105640522 | 0.58 |
ENSMUST00000106785.1
ENSMUST00000106786.1 ENSMUST00000106780.1 ENSMUST00000106784.1 |
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr17_-_25240112 | 0.57 |
ENSMUST00000038973.6
ENSMUST00000115154.4 |
Gnptg
|
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
| chr17_+_84956718 | 0.57 |
ENSMUST00000112305.3
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr4_-_149485157 | 0.57 |
ENSMUST00000126896.1
ENSMUST00000105693.1 ENSMUST00000030845.6 |
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr15_-_81360739 | 0.57 |
ENSMUST00000023040.7
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr2_-_84678051 | 0.57 |
ENSMUST00000053664.8
ENSMUST00000111664.1 |
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr2_-_156144138 | 0.56 |
ENSMUST00000109600.1
ENSMUST00000029147.9 |
Nfs1
|
nitrogen fixation gene 1 (S. cerevisiae) |
| chr5_+_34543365 | 0.55 |
ENSMUST00000101316.3
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr7_+_127841817 | 0.55 |
ENSMUST00000121705.1
|
Stx4a
|
syntaxin 4A (placental) |
| chr8_+_109778863 | 0.55 |
ENSMUST00000034171.8
|
Ap1g1
|
adaptor protein complex AP-1, gamma 1 subunit |
| chr14_+_54254124 | 0.55 |
ENSMUST00000180359.1
|
Abhd4
|
abhydrolase domain containing 4 |
| chr1_+_163929765 | 0.53 |
ENSMUST00000027876.4
ENSMUST00000170359.1 |
Scyl3
|
SCY1-like 3 (S. cerevisiae) |
| chr8_+_113635787 | 0.53 |
ENSMUST00000035777.8
|
Mon1b
|
MON1 homolog b (yeast) |
| chr7_-_126583177 | 0.53 |
ENSMUST00000098036.2
ENSMUST00000032962.4 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr14_+_55591708 | 0.53 |
ENSMUST00000019443.8
|
Rnf31
|
ring finger protein 31 |
| chr9_-_88438898 | 0.53 |
ENSMUST00000173011.1
ENSMUST00000174806.1 |
Snx14
|
sorting nexin 14 |
| chr6_+_29348069 | 0.53 |
ENSMUST00000173216.1
ENSMUST00000031779.10 ENSMUST00000090481.7 |
Calu
|
calumenin |
| chr6_-_72390659 | 0.53 |
ENSMUST00000059983.9
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr7_+_105640448 | 0.53 |
ENSMUST00000058333.3
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr5_+_113735782 | 0.53 |
ENSMUST00000065698.5
|
Ficd
|
FIC domain containing |
| chr1_+_128244122 | 0.53 |
ENSMUST00000027592.3
|
Ubxn4
|
UBX domain protein 4 |
| chr9_+_45055211 | 0.52 |
ENSMUST00000114663.2
|
Mpzl3
|
myelin protein zero-like 3 |
| chr19_+_46599081 | 0.52 |
ENSMUST00000138302.2
ENSMUST00000099376.4 |
Wbp1l
|
WW domain binding protein 1 like |
| chr1_-_13589717 | 0.52 |
ENSMUST00000027068.4
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr15_-_99820072 | 0.51 |
ENSMUST00000109024.2
|
Lima1
|
LIM domain and actin binding 1 |
| chr8_+_47713266 | 0.51 |
ENSMUST00000180928.1
|
E030037K01Rik
|
RIKEN cDNA E030037K01 gene |
| chr17_+_29268788 | 0.51 |
ENSMUST00000064709.5
ENSMUST00000120346.1 |
BC004004
|
cDNA sequence BC004004 |
| chr1_-_78196832 | 0.51 |
ENSMUST00000004994.9
|
Pax3
|
paired box gene 3 |
| chr9_+_45055166 | 0.51 |
ENSMUST00000114664.1
ENSMUST00000093856.3 |
Mpzl3
|
myelin protein zero-like 3 |
| chrX_+_107089234 | 0.50 |
ENSMUST00000118666.1
ENSMUST00000053375.3 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr15_+_76671615 | 0.50 |
ENSMUST00000037551.8
|
Ppp1r16a
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A |
| chr2_-_12419387 | 0.50 |
ENSMUST00000124515.1
|
Fam188a
|
family with sequence similarity 188, member A |
| chr17_+_25136622 | 0.50 |
ENSMUST00000160961.1
|
Clcn7
|
chloride channel 7 |
| chr13_-_9878998 | 0.50 |
ENSMUST00000063093.9
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr16_-_56717446 | 0.50 |
ENSMUST00000065515.7
|
Tfg
|
Trk-fused gene |
| chr7_+_127841752 | 0.49 |
ENSMUST00000033075.7
|
Stx4a
|
syntaxin 4A (placental) |
| chr7_+_27591705 | 0.48 |
ENSMUST00000167435.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr11_-_119355484 | 0.48 |
ENSMUST00000100172.2
ENSMUST00000005173.4 |
Sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr3_+_135825648 | 0.48 |
ENSMUST00000180196.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr9_-_58370369 | 0.48 |
ENSMUST00000040217.4
|
Tbc1d21
|
TBC1 domain family, member 21 |
| chr13_+_77135513 | 0.48 |
ENSMUST00000168779.1
|
2210408I21Rik
|
RIKEN cDNA 2210408I21 gene |
| chr9_+_46273064 | 0.48 |
ENSMUST00000156440.1
ENSMUST00000034583.6 ENSMUST00000114552.3 |
Zfp259
|
zinc finger protein 259 |
| chr10_+_39899304 | 0.47 |
ENSMUST00000181590.1
|
4930547M16Rik
|
RIKEN cDNA 4930547M16 gene |
| chr19_+_46623387 | 0.47 |
ENSMUST00000111855.4
|
Wbp1l
|
WW domain binding protein 1 like |
| chr9_-_88438940 | 0.47 |
ENSMUST00000165315.1
ENSMUST00000173039.1 |
Snx14
|
sorting nexin 14 |
| chr11_-_83302586 | 0.47 |
ENSMUST00000176374.1
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr1_+_58802492 | 0.47 |
ENSMUST00000165549.1
|
Casp8
|
caspase 8 |
| chr15_-_58933688 | 0.46 |
ENSMUST00000110155.1
|
Tatdn1
|
TatD DNase domain containing 1 |
| chr7_+_26757153 | 0.46 |
ENSMUST00000077855.6
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr9_+_122888471 | 0.46 |
ENSMUST00000063980.6
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr18_+_36783222 | 0.46 |
ENSMUST00000019287.8
|
Hars2
|
histidyl-tRNA synthetase 2, mitochondrial (putative) |
| chr16_-_56717286 | 0.46 |
ENSMUST00000121554.1
ENSMUST00000128551.1 |
Tfg
|
Trk-fused gene |
| chr2_-_127208274 | 0.45 |
ENSMUST00000056146.1
|
1810024B03Rik
|
RIKEN cDNA 1810024B03 gene |
| chr5_-_146220901 | 0.45 |
ENSMUST00000169407.2
ENSMUST00000161331.1 ENSMUST00000159074.2 ENSMUST00000067837.3 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr1_-_78197112 | 0.44 |
ENSMUST00000087086.6
|
Pax3
|
paired box gene 3 |
| chr3_+_116008220 | 0.44 |
ENSMUST00000106502.1
|
Extl2
|
exostoses (multiple)-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spdef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.6 | 1.7 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 1.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.4 | 1.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.4 | 1.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.3 | 1.0 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 1.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 1.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 7.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 2.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 1.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 0.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 0.9 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 | 0.8 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) release from viral latency(GO:0019046) |
| 0.3 | 0.8 | GO:1902022 | regulation of renal output by angiotensin(GO:0002019) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) L-lysine transport(GO:1902022) |
| 0.2 | 0.2 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.2 | 0.7 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.9 | GO:1902276 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.2 | 0.7 | GO:0060599 | cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.2 | 1.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 0.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.6 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 1.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 0.6 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 3.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.2 | 0.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 0.9 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.2 | 0.7 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of male germ cell proliferation(GO:2000256) |
| 0.2 | 1.7 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 2.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 3.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 3.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 2.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.4 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.3 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 1.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.4 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.6 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.5 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 2.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.2 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 1.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.8 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.5 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.6 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 1.6 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.1 | 0.8 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 0.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 1.6 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.9 | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 1.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.7 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 1.8 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 2.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 1.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 2.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 1.0 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.0 | 0.1 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 1.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.3 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.9 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 1.5 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0031119 | rRNA pseudouridine synthesis(GO:0031118) tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.8 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 1.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.4 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.3 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 1.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.2 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.8 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.3 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 1.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 3.8 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 1.2 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.9 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 1.0 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 0.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 1.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.9 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.3 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 6.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0036452 | ESCRT III complex(GO:0000815) ESCRT complex(GO:0036452) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.3 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 4.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.5 | 1.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 2.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 3.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 1.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.4 | 1.5 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.4 | 2.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 1.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 1.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.3 | 0.9 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.3 | 0.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 0.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 1.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 0.7 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 0.9 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 1.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 0.6 | GO:0004534 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.9 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.6 | GO:0031711 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.2 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 1.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.7 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 1.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 1.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.8 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.2 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 1.1 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0051381 | histamine binding(GO:0051381) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 5.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 5.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |