Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Spic
Z-value: 4.87
Transcription factors associated with Spic
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spic
|
ENSMUSG00000004359.10 | Spi-C transcription factor (Spi-1/PU.1 related) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spic | mm10_v2_chr10_-_88683021_88683025 | 0.93 | 1.2e-16 | Click! |
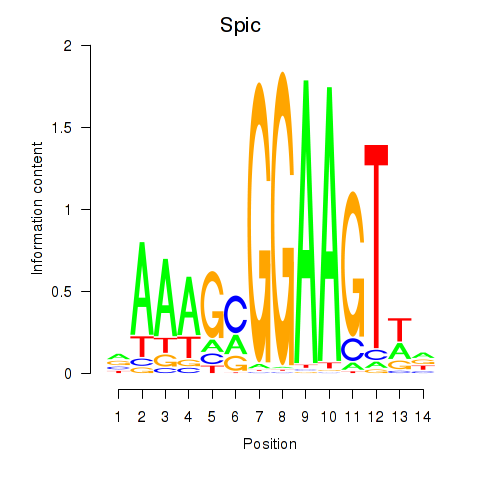
Activity profile of Spic motif
Sorted Z-values of Spic motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_106167564 | 37.33 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chrX_-_7964166 | 33.57 |
ENSMUST00000128449.1
|
Gata1
|
GATA binding protein 1 |
| chr11_+_87793470 | 29.82 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr11_+_87793722 | 28.52 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr10_+_79879614 | 25.08 |
ENSMUST00000006679.8
|
Prtn3
|
proteinase 3 |
| chr14_+_80000292 | 23.89 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr9_+_7558429 | 19.80 |
ENSMUST00000018765.2
|
Mmp8
|
matrix metallopeptidase 8 |
| chr1_-_144775419 | 18.01 |
ENSMUST00000027603.3
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr8_+_70373541 | 17.24 |
ENSMUST00000003659.7
|
Comp
|
cartilage oligomeric matrix protein |
| chr7_-_122132844 | 16.16 |
ENSMUST00000106469.1
ENSMUST00000063587.6 ENSMUST00000106468.1 ENSMUST00000130149.1 ENSMUST00000098068.3 |
Palb2
|
partner and localizer of BRCA2 |
| chr7_-_100856289 | 15.47 |
ENSMUST00000139604.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chrX_-_49788204 | 14.90 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr1_+_40084764 | 14.67 |
ENSMUST00000027243.7
|
Il1r2
|
interleukin 1 receptor, type II |
| chrX_+_8271133 | 14.56 |
ENSMUST00000127103.1
ENSMUST00000115591.1 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr11_+_98907801 | 14.01 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr11_+_32276400 | 13.87 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr16_-_75909272 | 13.10 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr19_+_6084983 | 12.75 |
ENSMUST00000025704.2
|
Cdca5
|
cell division cycle associated 5 |
| chr7_-_126704736 | 12.45 |
ENSMUST00000131415.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_+_43408187 | 12.13 |
ENSMUST00000005592.6
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chrX_+_8271381 | 11.91 |
ENSMUST00000033512.4
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr13_-_110357136 | 11.90 |
ENSMUST00000058806.5
|
Gapt
|
Grb2-binding adaptor, transmembrane |
| chr7_-_126704522 | 11.38 |
ENSMUST00000135087.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr9_+_56089962 | 10.98 |
ENSMUST00000059206.7
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr5_-_138170992 | 10.93 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr2_+_152847993 | 10.60 |
ENSMUST00000028969.8
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr11_-_102469839 | 10.60 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chr2_+_152847961 | 10.42 |
ENSMUST00000164120.1
ENSMUST00000178997.1 ENSMUST00000109816.1 |
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr7_+_28982832 | 10.40 |
ENSMUST00000085835.6
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr5_-_138171248 | 10.30 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr7_-_126704179 | 10.26 |
ENSMUST00000106364.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_+_128062657 | 10.19 |
ENSMUST00000120355.1
ENSMUST00000106240.2 ENSMUST00000098015.3 |
Itgam
|
integrin alpha M |
| chr7_-_127042420 | 10.07 |
ENSMUST00000032915.6
|
Kif22
|
kinesin family member 22 |
| chr15_+_78244781 | 9.94 |
ENSMUST00000096357.5
ENSMUST00000133618.1 |
Ncf4
|
neutrophil cytosolic factor 4 |
| chr11_+_120948480 | 9.85 |
ENSMUST00000070653.6
|
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr5_+_149265035 | 9.76 |
ENSMUST00000130144.1
ENSMUST00000071130.3 |
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr7_-_4725082 | 9.44 |
ENSMUST00000086363.4
ENSMUST00000086364.4 |
Tmem150b
|
transmembrane protein 150B |
| chr5_-_138171216 | 9.27 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr5_-_134229581 | 9.23 |
ENSMUST00000111275.1
ENSMUST00000016094.6 ENSMUST00000144086.1 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr10_+_83722865 | 9.21 |
ENSMUST00000150459.1
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr14_+_55765956 | 9.08 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr13_+_30749226 | 8.85 |
ENSMUST00000021784.2
ENSMUST00000110307.1 |
Irf4
|
interferon regulatory factor 4 |
| chr6_-_136941694 | 8.72 |
ENSMUST00000032344.5
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr5_-_116024452 | 8.66 |
ENSMUST00000031486.7
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr10_+_75943020 | 8.65 |
ENSMUST00000121151.1
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr3_-_105932664 | 8.58 |
ENSMUST00000098758.2
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr10_-_117282262 | 8.51 |
ENSMUST00000092163.7
|
Lyz2
|
lysozyme 2 |
| chr10_+_115817247 | 8.48 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr2_-_165400398 | 8.35 |
ENSMUST00000029213.4
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr7_+_142460834 | 8.09 |
ENSMUST00000018963.4
ENSMUST00000105967.1 |
Lsp1
|
lymphocyte specific 1 |
| chr3_+_103832562 | 7.91 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr4_+_44300876 | 7.90 |
ENSMUST00000045607.5
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr5_-_116024475 | 7.90 |
ENSMUST00000111999.1
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr7_-_45239041 | 7.89 |
ENSMUST00000131290.1
|
Cd37
|
CD37 antigen |
| chr7_+_128062698 | 7.82 |
ENSMUST00000119696.1
|
Itgam
|
integrin alpha M |
| chr7_-_45238794 | 7.75 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chr7_+_142460809 | 7.68 |
ENSMUST00000105968.1
|
Lsp1
|
lymphocyte specific 1 |
| chr8_+_84701430 | 7.66 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr9_-_114781986 | 7.62 |
ENSMUST00000035009.8
ENSMUST00000084867.7 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr10_-_30655859 | 7.62 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr17_+_7925990 | 7.58 |
ENSMUST00000036370.7
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr11_-_79523760 | 7.32 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr17_+_34604262 | 7.31 |
ENSMUST00000174041.1
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr4_-_133874682 | 7.07 |
ENSMUST00000168974.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_-_17008647 | 7.03 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chrX_+_20703906 | 7.02 |
ENSMUST00000033383.2
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr2_+_22774081 | 6.95 |
ENSMUST00000014290.8
|
Apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr15_-_78305603 | 6.95 |
ENSMUST00000096356.3
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr7_-_45239108 | 6.93 |
ENSMUST00000033063.6
|
Cd37
|
CD37 antigen |
| chr8_-_122432924 | 6.87 |
ENSMUST00000017604.8
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr1_+_87620334 | 6.62 |
ENSMUST00000042275.8
ENSMUST00000168783.1 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr2_+_24345282 | 6.62 |
ENSMUST00000114485.2
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr1_+_87620306 | 6.40 |
ENSMUST00000169754.1
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr6_-_136941887 | 6.37 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_+_24345305 | 6.33 |
ENSMUST00000114482.1
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr7_-_134232125 | 6.32 |
ENSMUST00000127524.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr2_+_156840966 | 6.30 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr7_-_134232005 | 6.29 |
ENSMUST00000134504.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr17_+_57279094 | 6.16 |
ENSMUST00000169220.2
ENSMUST00000005889.9 ENSMUST00000112870.4 |
Vav1
|
vav 1 oncogene |
| chr11_-_115133981 | 6.15 |
ENSMUST00000106561.1
ENSMUST00000051264.7 ENSMUST00000106562.2 |
Cd300lf
|
CD300 antigen like family member F |
| chr17_-_6655939 | 6.13 |
ENSMUST00000179554.1
|
Dynlt1f
|
dynein light chain Tctex-type 1F |
| chr19_+_53529100 | 6.08 |
ENSMUST00000038287.6
|
Dusp5
|
dual specificity phosphatase 5 |
| chr4_+_11758147 | 6.01 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chr17_-_6317474 | 5.99 |
ENSMUST00000169415.1
|
Dynlt1a
|
dynein light chain Tctex-type 1A |
| chr2_+_164805082 | 5.96 |
ENSMUST00000052107.4
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr5_+_34999111 | 5.92 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr2_+_43748802 | 5.87 |
ENSMUST00000112824.1
ENSMUST00000055776.7 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr9_-_20644726 | 5.82 |
ENSMUST00000148631.1
ENSMUST00000131128.1 ENSMUST00000151861.1 ENSMUST00000131343.1 ENSMUST00000086458.3 |
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr5_+_4192367 | 5.70 |
ENSMUST00000177258.1
|
Gm9897
|
predicted gene 9897 |
| chr5_+_34999046 | 5.69 |
ENSMUST00000114281.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr8_+_117498272 | 5.65 |
ENSMUST00000081232.7
|
Plcg2
|
phospholipase C, gamma 2 |
| chr10_+_130322845 | 5.64 |
ENSMUST00000042586.8
|
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr6_-_136941494 | 5.59 |
ENSMUST00000111892.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_+_36216749 | 5.48 |
ENSMUST00000147012.1
ENSMUST00000122948.1 |
Gm13431
|
predicted gene 13431 |
| chr12_-_32208470 | 5.43 |
ENSMUST00000085469.5
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr2_+_120463566 | 5.42 |
ENSMUST00000028749.7
ENSMUST00000110721.1 |
Capn3
|
calpain 3 |
| chr5_-_3893907 | 5.41 |
ENSMUST00000117463.1
ENSMUST00000044746.4 |
Mterf
|
mitochondrial transcription termination factor |
| chr4_+_130913264 | 5.39 |
ENSMUST00000156225.1
ENSMUST00000156742.1 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr14_+_116925516 | 5.39 |
ENSMUST00000125435.1
|
Gpc6
|
glypican 6 |
| chr1_-_171234290 | 5.16 |
ENSMUST00000079957.6
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr7_-_19271797 | 5.15 |
ENSMUST00000032561.8
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr1_-_170927567 | 5.14 |
ENSMUST00000046322.7
ENSMUST00000159171.1 |
Fcrla
|
Fc receptor-like A |
| chr3_-_67463828 | 5.09 |
ENSMUST00000058981.2
|
Lxn
|
latexin |
| chr12_-_11265768 | 5.03 |
ENSMUST00000166117.1
|
Gen1
|
Gen homolog 1, endonuclease (Drosophila) |
| chr14_+_57524734 | 5.03 |
ENSMUST00000089494.4
|
Il17d
|
interleukin 17D |
| chr1_-_149961230 | 5.02 |
ENSMUST00000070200.8
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr19_+_4154606 | 4.99 |
ENSMUST00000061086.8
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr2_+_91650116 | 4.99 |
ENSMUST00000111331.2
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr15_+_103453782 | 4.93 |
ENSMUST00000047405.7
|
Nckap1l
|
NCK associated protein 1 like |
| chr7_+_28440927 | 4.90 |
ENSMUST00000078845.6
|
Gmfg
|
glia maturation factor, gamma |
| chr1_+_152807877 | 4.80 |
ENSMUST00000027754.6
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chrX_-_136085206 | 4.80 |
ENSMUST00000138878.1
ENSMUST00000080929.6 |
Nxf3
|
nuclear RNA export factor 3 |
| chr1_-_20820213 | 4.72 |
ENSMUST00000053266.9
|
Mcm3
|
minichromosome maintenance deficient 3 (S. cerevisiae) |
| chr12_-_78980758 | 4.70 |
ENSMUST00000174072.1
|
Tmem229b
|
transmembrane protein 229B |
| chr15_-_66812593 | 4.70 |
ENSMUST00000100572.3
|
Sla
|
src-like adaptor |
| chr7_+_126584937 | 4.66 |
ENSMUST00000039522.6
|
Apobr
|
apolipoprotein B receptor |
| chr3_-_101029543 | 4.62 |
ENSMUST00000147399.2
|
Cd101
|
CD101 antigen |
| chr17_+_47594629 | 4.61 |
ENSMUST00000182846.1
|
Ccnd3
|
cyclin D3 |
| chr6_-_124733067 | 4.60 |
ENSMUST00000173647.1
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_-_170927540 | 4.59 |
ENSMUST00000162136.1
ENSMUST00000162887.1 |
Fcrla
|
Fc receptor-like A |
| chr12_-_32208609 | 4.58 |
ENSMUST00000053215.7
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| chr2_+_164074122 | 4.57 |
ENSMUST00000018353.7
|
Stk4
|
serine/threonine kinase 4 |
| chr2_-_105399286 | 4.54 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr6_+_123229843 | 4.52 |
ENSMUST00000112554.2
ENSMUST00000024118.4 ENSMUST00000117130.1 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr17_+_29360923 | 4.50 |
ENSMUST00000024810.6
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr9_-_77045788 | 4.50 |
ENSMUST00000034911.6
|
Tinag
|
tubulointerstitial nephritis antigen |
| chr9_+_109875541 | 4.47 |
ENSMUST00000094324.3
|
Cdc25a
|
cell division cycle 25A |
| chr11_+_70505244 | 4.44 |
ENSMUST00000019063.2
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr3_+_88621102 | 4.42 |
ENSMUST00000029694.7
ENSMUST00000176804.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr4_+_115839192 | 4.38 |
ENSMUST00000019677.5
ENSMUST00000144427.1 ENSMUST00000106513.3 ENSMUST00000130819.1 ENSMUST00000151203.1 ENSMUST00000140315.1 |
Mknk1
|
MAP kinase-interacting serine/threonine kinase 1 |
| chr17_-_33955658 | 4.37 |
ENSMUST00000174609.2
ENSMUST00000008812.7 |
Rps18
|
ribosomal protein S18 |
| chr4_+_130913120 | 4.25 |
ENSMUST00000151698.1
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr2_+_91650169 | 4.23 |
ENSMUST00000090614.4
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr13_-_97137877 | 4.21 |
ENSMUST00000073456.7
|
Nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr3_+_87376381 | 4.20 |
ENSMUST00000163661.1
ENSMUST00000072480.2 ENSMUST00000167200.1 |
Fcrl1
|
Fc receptor-like 1 |
| chr6_+_5725639 | 4.12 |
ENSMUST00000115556.1
ENSMUST00000115555.1 ENSMUST00000115559.3 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr5_-_99978914 | 4.00 |
ENSMUST00000112939.3
ENSMUST00000171786.1 ENSMUST00000072750.6 ENSMUST00000019128.8 ENSMUST00000172361.1 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr17_+_48316141 | 4.00 |
ENSMUST00000049614.6
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr9_+_106222598 | 4.00 |
ENSMUST00000062241.9
|
Tlr9
|
toll-like receptor 9 |
| chr11_+_69914179 | 3.99 |
ENSMUST00000057884.5
|
Gps2
|
G protein pathway suppressor 2 |
| chr13_-_49309217 | 3.96 |
ENSMUST00000110087.2
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr4_+_140701466 | 3.91 |
ENSMUST00000038893.5
ENSMUST00000138808.1 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr3_+_109123104 | 3.89 |
ENSMUST00000029477.6
|
Slc25a24
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 |
| chr4_-_41314877 | 3.89 |
ENSMUST00000030145.8
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr5_+_34999070 | 3.88 |
ENSMUST00000114280.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr3_+_105904377 | 3.86 |
ENSMUST00000000574.1
|
Adora3
|
adenosine A3 receptor |
| chr2_+_119112793 | 3.84 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr12_+_98268626 | 3.77 |
ENSMUST00000075072.4
|
Gpr65
|
G-protein coupled receptor 65 |
| chr11_+_70130329 | 3.76 |
ENSMUST00000041550.5
ENSMUST00000165951.1 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chr1_-_33669745 | 3.76 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr1_-_37496095 | 3.71 |
ENSMUST00000148047.1
ENSMUST00000143636.1 |
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr7_-_35396708 | 3.67 |
ENSMUST00000154597.1
ENSMUST00000032704.5 |
C230052I12Rik
|
RIKEN cDNA C230052I12 gene |
| chr17_+_6601671 | 3.66 |
ENSMUST00000092966.4
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr6_-_67376147 | 3.60 |
ENSMUST00000018485.3
|
Il12rb2
|
interleukin 12 receptor, beta 2 |
| chr18_+_67562387 | 3.57 |
ENSMUST00000163749.1
|
Gm17669
|
predicted gene, 17669 |
| chr7_-_104369782 | 3.56 |
ENSMUST00000164410.1
|
Trim30b
|
tripartite motif-containing 30B |
| chr9_+_108508005 | 3.56 |
ENSMUST00000006838.8
ENSMUST00000134939.1 |
Qars
|
glutaminyl-tRNA synthetase |
| chr1_+_179546303 | 3.53 |
ENSMUST00000040706.8
|
Cnst
|
consortin, connexin sorting protein |
| chr5_-_38876693 | 3.50 |
ENSMUST00000169819.1
ENSMUST00000171633.1 |
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr1_-_52232296 | 3.48 |
ENSMUST00000114512.1
|
Gls
|
glutaminase |
| chr7_-_19604444 | 3.47 |
ENSMUST00000086041.5
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr19_+_4878671 | 3.44 |
ENSMUST00000006632.7
|
Zdhhc24
|
zinc finger, DHHC domain containing 24 |
| chr3_+_24333046 | 3.38 |
ENSMUST00000077389.6
|
Gm7536
|
predicted gene 7536 |
| chr5_-_137116177 | 3.35 |
ENSMUST00000054384.5
ENSMUST00000152207.1 |
Trim56
|
tripartite motif-containing 56 |
| chr4_-_117891994 | 3.27 |
ENSMUST00000030265.3
|
Dph2
|
DPH2 homolog (S. cerevisiae) |
| chr1_+_191325912 | 3.27 |
ENSMUST00000027940.5
|
Tmem206
|
transmembrane protein 206 |
| chr16_+_91406235 | 3.25 |
ENSMUST00000023691.5
|
Il10rb
|
interleukin 10 receptor, beta |
| chr8_+_94923687 | 3.19 |
ENSMUST00000153448.1
ENSMUST00000074570.3 ENSMUST00000166802.1 |
Gpr114
|
G protein-coupled receptor 114 |
| chr8_-_70897407 | 3.18 |
ENSMUST00000054220.8
|
Rpl18a
|
ribosomal protein L18A |
| chr2_-_91649785 | 3.12 |
ENSMUST00000111333.1
|
Zfp408
|
zinc finger protein 408 |
| chr11_-_69323768 | 3.10 |
ENSMUST00000092973.5
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr5_+_108065742 | 3.08 |
ENSMUST00000081567.4
ENSMUST00000170319.1 ENSMUST00000112626.1 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr5_+_86071734 | 3.06 |
ENSMUST00000031171.7
|
Stap1
|
signal transducing adaptor family member 1 |
| chr14_+_30479565 | 3.05 |
ENSMUST00000022535.7
|
Dcp1a
|
DCP1 decapping enzyme homolog A (S. cerevisiae) |
| chr7_+_131032061 | 3.04 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr12_-_24493656 | 2.97 |
ENSMUST00000073088.2
|
Gm16372
|
predicted pseudogene 16372 |
| chr1_+_136683375 | 2.96 |
ENSMUST00000181524.1
|
Gm19705
|
predicted gene, 19705 |
| chr12_+_77238093 | 2.95 |
ENSMUST00000177595.1
ENSMUST00000171770.2 |
Fut8
|
fucosyltransferase 8 |
| chr1_-_171059390 | 2.93 |
ENSMUST00000164044.1
ENSMUST00000169017.1 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr7_-_100964371 | 2.92 |
ENSMUST00000060174.4
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_+_35135463 | 2.89 |
ENSMUST00000173535.1
ENSMUST00000173952.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr10_-_89443888 | 2.87 |
ENSMUST00000099374.2
ENSMUST00000105298.1 |
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr11_+_120458093 | 2.84 |
ENSMUST00000058370.7
ENSMUST00000175970.1 ENSMUST00000176120.1 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr16_-_36642742 | 2.82 |
ENSMUST00000135280.1
|
Cd86
|
CD86 antigen |
| chr11_-_98400453 | 2.80 |
ENSMUST00000090827.5
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr10_-_44458687 | 2.77 |
ENSMUST00000105490.2
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr15_+_39745926 | 2.77 |
ENSMUST00000022913.4
|
Dcstamp
|
dentrocyte expressed seven transmembrane protein |
| chr19_-_56389877 | 2.75 |
ENSMUST00000166203.1
ENSMUST00000167239.1 ENSMUST00000040711.8 ENSMUST00000095947.4 ENSMUST00000073536.6 |
Nrap
|
nebulin-related anchoring protein |
| chr10_-_80102653 | 2.73 |
ENSMUST00000042771.7
|
Sbno2
|
strawberry notch homolog 2 (Drosophila) |
| chr7_+_28540863 | 2.71 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr2_-_91649751 | 2.71 |
ENSMUST00000099714.3
|
Zfp408
|
zinc finger protein 408 |
| chr8_+_94666722 | 2.67 |
ENSMUST00000034228.8
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr19_-_40994133 | 2.64 |
ENSMUST00000117695.1
|
Blnk
|
B cell linker |
| chr2_-_84670727 | 2.63 |
ENSMUST00000117299.2
|
2700094K13Rik
|
RIKEN cDNA 2700094K13 gene |
| chr11_-_98400393 | 2.63 |
ENSMUST00000128897.1
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr1_-_150164943 | 2.62 |
ENSMUST00000181308.1
|
Gm26687
|
predicted gene, 26687 |
| chr2_-_167492826 | 2.61 |
ENSMUST00000109211.2
ENSMUST00000057627.9 |
Spata2
|
spermatogenesis associated 2 |
| chr11_-_102880981 | 2.59 |
ENSMUST00000107060.1
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr6_+_137410721 | 2.55 |
ENSMUST00000167002.1
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr2_-_93046053 | 2.53 |
ENSMUST00000111272.1
ENSMUST00000178666.1 ENSMUST00000147339.1 |
Prdm11
|
PR domain containing 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spic
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.4 | 58.3 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 11.2 | 33.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 5.7 | 34.1 | GO:0032796 | uropod organization(GO:0032796) |
| 5.5 | 27.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 5.0 | 25.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 4.2 | 12.7 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 3.5 | 14.0 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 3.4 | 34.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 3.3 | 13.0 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 3.1 | 15.5 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 2.9 | 8.8 | GO:0045404 | interleukin-10 biosynthetic process(GO:0042091) interleukin-13 biosynthetic process(GO:0042231) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 2.8 | 11.1 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 2.6 | 20.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 2.6 | 18.0 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 2.5 | 7.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 2.4 | 9.8 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 2.4 | 9.5 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 2.3 | 6.9 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 2.3 | 22.6 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 2.1 | 6.2 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 2.0 | 6.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 2.0 | 4.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 1.9 | 5.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 1.8 | 5.4 | GO:0070315 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.7 | 13.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.7 | 5.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 1.6 | 4.9 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.6 | 4.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 1.5 | 9.2 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 1.5 | 26.5 | GO:0015816 | glycine transport(GO:0015816) |
| 1.4 | 10.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 1.4 | 7.0 | GO:0070560 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 1.4 | 4.2 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 1.4 | 15.3 | GO:0035878 | nail development(GO:0035878) |
| 1.4 | 4.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 1.2 | 4.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.2 | 3.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 1.2 | 3.5 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 1.2 | 5.8 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.2 | 6.9 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 1.2 | 6.9 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.2 | 4.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.1 | 4.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.1 | 5.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 1.1 | 7.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 1.1 | 8.6 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 1.0 | 3.1 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 1.0 | 5.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 1.0 | 3.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 1.0 | 3.0 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 1.0 | 18.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.0 | 9.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.9 | 2.8 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.9 | 3.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.9 | 1.8 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.9 | 8.7 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.8 | 5.0 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.8 | 3.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.8 | 17.5 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.8 | 7.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.8 | 3.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.7 | 2.9 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.7 | 5.0 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.7 | 4.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.6 | 3.9 | GO:0015867 | ATP transport(GO:0015867) |
| 0.6 | 7.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 1.9 | GO:1904732 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.6 | 8.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 1.8 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.6 | 3.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 4.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.5 | 13.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.5 | 2.7 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.5 | 13.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.5 | 1.6 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.5 | 7.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.5 | 4.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 2.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.5 | 10.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 0.9 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.5 | 2.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.4 | 2.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.4 | 3.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.4 | 23.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.4 | 3.9 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.4 | 0.9 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 2.5 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.4 | 0.8 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.4 | 6.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 9.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.4 | 3.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 4.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 12.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.4 | 2.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.4 | 1.4 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 | 1.0 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.3 | 1.7 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 1.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 1.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.3 | 1.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.3 | 0.7 | GO:0035419 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.3 | 1.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.6 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 | 0.9 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 1.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 1.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.3 | 6.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.3 | 1.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 1.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.7 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 1.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.3 | 4.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 4.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 17.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.2 | 5.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 1.7 | GO:1904851 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 2.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 5.0 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 4.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 1.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 2.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 4.0 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.2 | 4.5 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 3.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 1.7 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.2 | 3.8 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 0.2 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.2 | 4.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.5 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 4.9 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 8.5 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.8 | GO:0045651 | positive regulation of granulocyte differentiation(GO:0030854) positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 1.6 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 1.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 3.3 | GO:0051764 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 0.1 | 9.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.6 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 1.7 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 0.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 5.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.5 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 6.1 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 | 0.7 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 1.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 3.2 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 12.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 3.7 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.1 | 2.5 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 2.0 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 1.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 4.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 2.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.8 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 0.3 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 1.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 4.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 2.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.7 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 3.6 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.1 | 8.9 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 4.9 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 1.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 6.3 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 6.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.9 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 4.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 9.3 | GO:0006935 | chemotaxis(GO:0006935) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 5.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 3.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 2.4 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.7 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 58.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 2.9 | 11.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 2.8 | 30.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 2.6 | 21.0 | GO:0005818 | aster(GO:0005818) |
| 2.5 | 25.4 | GO:0042581 | specific granule(GO:0042581) |
| 2.3 | 35.2 | GO:0042555 | MCM complex(GO:0042555) |
| 2.1 | 2.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 1.7 | 13.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.6 | 4.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 1.5 | 9.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.5 | 34.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.4 | 8.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.4 | 12.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 1.4 | 4.1 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.4 | 11.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.2 | 8.6 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.1 | 3.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 1.0 | 4.0 | GO:0036019 | endolysosome(GO:0036019) |
| 0.8 | 4.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.8 | 3.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.8 | 3.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.7 | 6.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.7 | 29.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.6 | 5.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 19.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 2.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 1.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.5 | 15.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.5 | 2.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 20.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 12.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 4.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 1.5 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.3 | 38.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 13.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 4.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 3.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 0.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 12.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 8.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 4.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 2.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 2.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 14.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 3.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 3.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 2.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 3.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 1.0 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 5.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 6.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 3.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 68.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 7.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 4.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 5.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 3.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 11.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 4.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 19.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 1.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 5.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 63.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 5.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 6.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 3.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 8.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 38.7 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 4.3 | 12.9 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) interleukin-1 Type I receptor antagonist activity(GO:0045352) interleukin-1 Type II receptor antagonist activity(GO:0045353) |
| 3.3 | 9.8 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 2.7 | 24.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 2.6 | 20.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.6 | 18.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 2.5 | 17.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 2.3 | 21.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 2.3 | 9.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 2.2 | 13.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 2.1 | 10.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 2.1 | 33.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 2.1 | 6.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 2.0 | 6.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.9 | 26.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 1.7 | 13.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.7 | 14.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.5 | 34.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.5 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.4 | 9.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.3 | 5.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 1.0 | 3.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.0 | 2.9 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.9 | 10.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.9 | 7.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.9 | 54.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.9 | 6.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 2.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.8 | 4.7 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.7 | 29.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.7 | 10.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 5.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.7 | 5.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 8.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 3.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.6 | 2.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.6 | 5.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 3.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 12.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 8.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 3.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.5 | 15.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.5 | 3.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 3.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.4 | 4.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 1.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 4.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 6.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 4.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 5.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 24.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.4 | 3.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 7.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 5.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 3.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 5.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 17.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 3.9 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 6.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 3.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 3.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 1.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 4.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 1.7 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 12.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 7.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 2.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 4.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 43.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 7.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.6 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 1.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.2 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 7.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 0.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 8.8 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 0.8 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 4.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 45.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 5.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 4.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 4.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 3.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 5.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.9 | GO:0043142 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 7.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 3.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.6 | GO:0008823 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 8.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 4.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 4.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 17.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 2.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 3.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 5.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 26.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 4.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 4.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 26.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 2.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 6.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 1.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 59.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 1.1 | 53.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.9 | 29.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.8 | 12.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 24.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.5 | 15.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 16.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.5 | 20.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 15.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.5 | 32.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.4 | 15.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 15.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.4 | 10.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 17.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 24.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 15.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 3.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 5.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 4.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 22.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 8.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 4.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 10.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 2.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 8.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 2.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 4.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 5.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 26.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 9.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 5.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 4.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 8.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 5.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 4.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 35.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.5 | 4.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 1.3 | 14.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.0 | 17.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.8 | 11.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.8 | 12.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 9.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 16.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.6 | 26.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 28.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.5 | 2.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.4 | 5.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 16.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.4 | 5.8 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 11.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 3.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 4.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 7.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 9.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.3 | 16.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.3 | 8.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 4.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 7.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 2.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 53.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 7.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 4.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 14.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 3.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 30.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 6.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 16.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 3.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 12.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 2.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 6.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 4.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 4.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 7.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 5.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 4.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 31.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 5.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 2.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 7.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 6.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 6.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 6.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 4.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |