Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
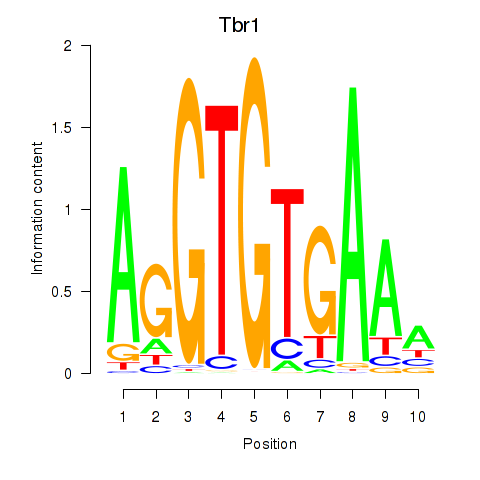
Results for Tbr1
Z-value: 1.02
Transcription factors associated with Tbr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbr1
|
ENSMUSG00000035033.9 | T-box brain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbr1 | mm10_v2_chr2_+_61804453_61804538 | 0.15 | 3.8e-01 | Click! |
Activity profile of Tbr1 motif
Sorted Z-values of Tbr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_93654863 | 2.90 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr14_+_80000292 | 2.69 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr14_-_51057242 | 2.42 |
ENSMUST00000089798.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr2_+_84980458 | 2.20 |
ENSMUST00000028467.5
|
Prg2
|
proteoglycan 2, bone marrow |
| chr5_-_108749448 | 2.19 |
ENSMUST00000068946.7
|
Rnf212
|
ring finger protein 212 |
| chr10_+_75573448 | 2.00 |
ENSMUST00000006508.3
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr16_+_32756336 | 2.00 |
ENSMUST00000135753.1
|
Muc4
|
mucin 4 |
| chr1_-_132390301 | 1.95 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr17_+_29114142 | 1.92 |
ENSMUST00000141797.1
ENSMUST00000132262.1 ENSMUST00000141239.1 ENSMUST00000138816.1 |
Gm16194
|
predicted gene 16194 |
| chr9_+_98490522 | 1.83 |
ENSMUST00000035029.2
|
Rbp2
|
retinol binding protein 2, cellular |
| chr12_+_109459843 | 1.83 |
ENSMUST00000173812.1
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr19_-_11640828 | 1.82 |
ENSMUST00000112984.2
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr11_+_70639118 | 1.76 |
ENSMUST00000055184.6
ENSMUST00000108551.2 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chrX_+_136138996 | 1.75 |
ENSMUST00000116527.1
|
Bex4
|
brain expressed gene 4 |
| chr2_+_148798785 | 1.72 |
ENSMUST00000028931.3
ENSMUST00000109947.1 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr3_+_153844209 | 1.58 |
ENSMUST00000044089.3
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr12_-_78906929 | 1.53 |
ENSMUST00000021544.7
|
Plek2
|
pleckstrin 2 |
| chr11_-_69900930 | 1.51 |
ENSMUST00000018714.6
ENSMUST00000128046.1 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_-_69900949 | 1.51 |
ENSMUST00000102580.3
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr7_-_6730412 | 1.50 |
ENSMUST00000051209.4
|
Peg3
|
paternally expressed 3 |
| chr10_-_75940633 | 1.46 |
ENSMUST00000059658.4
|
Gm867
|
predicted gene 867 |
| chr7_-_24760311 | 1.46 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chr7_-_120982260 | 1.45 |
ENSMUST00000033169.8
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr11_+_32283511 | 1.43 |
ENSMUST00000093209.3
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr9_-_123678782 | 1.43 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr15_+_44457522 | 1.42 |
ENSMUST00000166957.1
ENSMUST00000038336.5 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr15_+_79891631 | 1.41 |
ENSMUST00000177350.1
ENSMUST00000177483.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr18_-_67549173 | 1.40 |
ENSMUST00000115050.1
|
Spire1
|
spire homolog 1 (Drosophila) |
| chr11_+_32296489 | 1.39 |
ENSMUST00000093207.3
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr15_+_80623499 | 1.34 |
ENSMUST00000043149.7
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr10_+_20347788 | 1.32 |
ENSMUST00000169712.1
|
Mtfr2
|
mitochondrial fission regulator 2 |
| chrX_+_100625737 | 1.31 |
ENSMUST00000048962.3
|
Kif4
|
kinesin family member 4 |
| chr7_+_110777653 | 1.29 |
ENSMUST00000148292.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr19_-_17356631 | 1.28 |
ENSMUST00000174236.1
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr7_-_141443314 | 1.25 |
ENSMUST00000106005.2
|
Lrdd
|
leucine-rich and death domain containing |
| chr4_-_129440800 | 1.22 |
ENSMUST00000053042.5
ENSMUST00000106046.1 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr14_-_60197173 | 1.21 |
ENSMUST00000131670.1
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr1_+_174158722 | 1.20 |
ENSMUST00000068403.3
|
Olfr420
|
olfactory receptor 420 |
| chr2_+_122637844 | 1.19 |
ENSMUST00000047498.8
|
AA467197
|
expressed sequence AA467197 |
| chr5_-_122002340 | 1.17 |
ENSMUST00000134326.1
|
Cux2
|
cut-like homeobox 2 |
| chr2_+_103970115 | 1.15 |
ENSMUST00000111143.1
ENSMUST00000138815.1 |
Lmo2
|
LIM domain only 2 |
| chr15_-_66801577 | 1.11 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr15_-_101491509 | 1.10 |
ENSMUST00000023718.7
|
5430421N21Rik
|
RIKEN cDNA 5430421N21 gene |
| chr17_+_48232755 | 1.10 |
ENSMUST00000113251.3
ENSMUST00000048782.6 |
Trem1
|
triggering receptor expressed on myeloid cells 1 |
| chr17_+_48247759 | 1.10 |
ENSMUST00000048065.5
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr12_+_109545390 | 1.08 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr16_-_20425881 | 1.05 |
ENSMUST00000077867.3
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr13_-_95444827 | 1.03 |
ENSMUST00000045583.7
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr1_-_133753681 | 1.03 |
ENSMUST00000125659.1
ENSMUST00000165602.2 ENSMUST00000048953.7 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr12_+_109743787 | 1.02 |
ENSMUST00000183068.1
|
Mirg
|
miRNA containing gene |
| chr7_-_3915501 | 1.01 |
ENSMUST00000038176.8
ENSMUST00000090689.4 |
Lilra6
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 |
| chr6_-_131313827 | 1.00 |
ENSMUST00000049150.1
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr6_-_122609964 | 1.00 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr14_-_89898466 | 0.99 |
ENSMUST00000081204.4
|
Gm10110
|
predicted gene 10110 |
| chr19_-_10203880 | 0.99 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr3_+_135212557 | 0.97 |
ENSMUST00000062893.7
|
Cenpe
|
centromere protein E |
| chr9_+_22411515 | 0.97 |
ENSMUST00000058868.7
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr7_-_30823766 | 0.95 |
ENSMUST00000053156.3
|
Ffar2
|
free fatty acid receptor 2 |
| chr16_-_88751628 | 0.94 |
ENSMUST00000053149.2
|
Krtap13
|
keratin associated protein 13 |
| chr9_+_108991902 | 0.94 |
ENSMUST00000147989.1
ENSMUST00000051873.8 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr6_-_36811361 | 0.92 |
ENSMUST00000101534.1
|
Ptn
|
pleiotrophin |
| chr1_+_40515362 | 0.92 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chrX_+_96455359 | 0.91 |
ENSMUST00000033553.7
|
Heph
|
hephaestin |
| chr11_-_8973266 | 0.91 |
ENSMUST00000154153.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr5_+_64092925 | 0.91 |
ENSMUST00000087324.5
|
Pgm1
|
phosphoglucomutase 1 |
| chrX_-_8145713 | 0.90 |
ENSMUST00000115615.2
ENSMUST00000115616.1 ENSMUST00000115621.2 |
Rbm3
|
RNA binding motif protein 3 |
| chr11_-_79059872 | 0.90 |
ENSMUST00000141409.1
|
Ksr1
|
kinase suppressor of ras 1 |
| chr15_+_79892436 | 0.90 |
ENSMUST00000175752.1
ENSMUST00000176325.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr18_+_82475133 | 0.88 |
ENSMUST00000091789.4
ENSMUST00000114676.1 ENSMUST00000047865.7 |
Mbp
|
myelin basic protein |
| chr2_+_144033059 | 0.87 |
ENSMUST00000037722.2
ENSMUST00000110032.1 |
Banf2
|
barrier to autointegration factor 2 |
| chr6_+_134929089 | 0.87 |
ENSMUST00000183867.1
ENSMUST00000184991.1 ENSMUST00000183905.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr1_+_90953108 | 0.87 |
ENSMUST00000166281.1
|
Prlh
|
prolactin releasing hormone |
| chr7_-_126414855 | 0.86 |
ENSMUST00000032968.5
|
Cd19
|
CD19 antigen |
| chr7_-_14254870 | 0.86 |
ENSMUST00000184731.1
ENSMUST00000076576.6 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr1_-_189688074 | 0.85 |
ENSMUST00000171929.1
ENSMUST00000165962.1 |
Cenpf
|
centromere protein F |
| chr7_-_3720382 | 0.84 |
ENSMUST00000078451.6
|
Pirb
|
paired Ig-like receptor B |
| chr16_+_18127607 | 0.83 |
ENSMUST00000059589.5
|
Rtn4r
|
reticulon 4 receptor |
| chr14_-_56085214 | 0.83 |
ENSMUST00000015594.7
|
Mcpt8
|
mast cell protease 8 |
| chr10_+_62252325 | 0.83 |
ENSMUST00000020278.5
|
Tacr2
|
tachykinin receptor 2 |
| chr9_+_111004811 | 0.83 |
ENSMUST00000080872.4
|
Gm10030
|
predicted gene 10030 |
| chr16_+_45610380 | 0.82 |
ENSMUST00000161347.2
ENSMUST00000023339.4 |
Gcsam
|
germinal center associated, signaling and motility |
| chr19_+_53460610 | 0.81 |
ENSMUST00000180442.1
|
4833407H14Rik
|
RIKEN cDNA 4833407H14 gene |
| chr14_-_59142886 | 0.80 |
ENSMUST00000022548.3
ENSMUST00000162674.1 ENSMUST00000159858.1 ENSMUST00000162271.1 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr13_+_44729535 | 0.80 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr12_-_113307912 | 0.80 |
ENSMUST00000103418.1
|
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr2_-_13491900 | 0.79 |
ENSMUST00000091436.5
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chrX_+_134295225 | 0.78 |
ENSMUST00000037687.7
|
Tmem35
|
transmembrane protein 35 |
| chr7_-_132813799 | 0.78 |
ENSMUST00000097998.2
|
Fam53b
|
family with sequence similarity 53, member B |
| chrX_-_150657392 | 0.77 |
ENSMUST00000151403.2
ENSMUST00000087253.4 ENSMUST00000112709.1 ENSMUST00000163969.1 ENSMUST00000087258.3 |
Tro
|
trophinin |
| chr9_+_99243421 | 0.77 |
ENSMUST00000093795.3
|
Cep70
|
centrosomal protein 70 |
| chr11_-_69900886 | 0.77 |
ENSMUST00000108621.2
ENSMUST00000100969.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr9_-_39604124 | 0.76 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr13_-_47105790 | 0.76 |
ENSMUST00000129352.1
|
Dek
|
DEK oncogene (DNA binding) |
| chr5_-_148399901 | 0.76 |
ENSMUST00000048116.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_103356324 | 0.76 |
ENSMUST00000136491.2
ENSMUST00000107023.2 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr2_+_122637867 | 0.75 |
ENSMUST00000110512.3
|
AA467197
|
expressed sequence AA467197 |
| chr1_+_171917512 | 0.75 |
ENSMUST00000171330.1
|
Slamf6
|
SLAM family member 6 |
| chr4_+_117849193 | 0.75 |
ENSMUST00000132043.2
ENSMUST00000169990.1 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_141443989 | 0.74 |
ENSMUST00000026580.5
|
Lrdd
|
leucine-rich and death domain containing |
| chr8_-_68121527 | 0.74 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chrX_+_11302432 | 0.74 |
ENSMUST00000179428.1
|
Gm14474
|
predicted gene 14474 |
| chr4_-_42084291 | 0.73 |
ENSMUST00000177937.1
|
Gm21968
|
predicted gene, 21968 |
| chr9_-_72491939 | 0.73 |
ENSMUST00000185151.1
ENSMUST00000085358.5 ENSMUST00000184125.1 ENSMUST00000183574.1 ENSMUST00000184831.1 |
Tex9
|
testis expressed gene 9 |
| chr16_-_20426375 | 0.73 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr6_+_134929118 | 0.73 |
ENSMUST00000185152.1
ENSMUST00000184504.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr15_+_79892397 | 0.71 |
ENSMUST00000175714.1
ENSMUST00000109620.3 ENSMUST00000165537.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr9_+_22454290 | 0.70 |
ENSMUST00000168332.1
|
Gm17545
|
predicted gene, 17545 |
| chr7_+_128062657 | 0.70 |
ENSMUST00000120355.1
ENSMUST00000106240.2 ENSMUST00000098015.3 |
Itgam
|
integrin alpha M |
| chr13_-_70841790 | 0.70 |
ENSMUST00000080145.6
ENSMUST00000109694.2 |
Adamts16
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 16 |
| chr11_-_70220776 | 0.69 |
ENSMUST00000141290.1
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr12_+_51348265 | 0.68 |
ENSMUST00000119211.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr7_-_127993831 | 0.67 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr13_-_100316616 | 0.67 |
ENSMUST00000042220.2
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr11_+_69913888 | 0.67 |
ENSMUST00000072581.2
ENSMUST00000116358.1 |
Gps2
|
G protein pathway suppressor 2 |
| chr7_-_132813528 | 0.67 |
ENSMUST00000097999.2
|
Fam53b
|
family with sequence similarity 53, member B |
| chr2_+_103970221 | 0.66 |
ENSMUST00000111140.2
ENSMUST00000111139.2 |
Lmo2
|
LIM domain only 2 |
| chr15_-_83033294 | 0.66 |
ENSMUST00000100377.4
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chr12_-_69228167 | 0.66 |
ENSMUST00000021359.5
|
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr13_+_44729794 | 0.66 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr15_-_103310425 | 0.66 |
ENSMUST00000079824.4
|
Gpr84
|
G protein-coupled receptor 84 |
| chr4_-_127330799 | 0.66 |
ENSMUST00000046532.3
|
Gjb3
|
gap junction protein, beta 3 |
| chr9_-_49798905 | 0.66 |
ENSMUST00000114476.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr8_+_84723003 | 0.65 |
ENSMUST00000098571.4
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr8_-_4259257 | 0.64 |
ENSMUST00000053252.7
|
Ctxn1
|
cortexin 1 |
| chr4_-_11965699 | 0.64 |
ENSMUST00000108301.1
ENSMUST00000095144.3 ENSMUST00000108302.1 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr9_+_73044833 | 0.63 |
ENSMUST00000184146.1
ENSMUST00000034722.4 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr9_+_50856924 | 0.63 |
ENSMUST00000174628.1
ENSMUST00000034560.7 ENSMUST00000114437.2 ENSMUST00000175645.1 ENSMUST00000176349.1 ENSMUST00000176798.1 ENSMUST00000175640.1 |
Ppp2r1b
|
protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform |
| chr9_-_95845215 | 0.63 |
ENSMUST00000093800.2
|
Pls1
|
plastin 1 (I-isoform) |
| chrX_+_136666375 | 0.63 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr7_-_42578588 | 0.63 |
ENSMUST00000179470.1
|
Gm21028
|
predicted gene, 21028 |
| chr4_+_103143052 | 0.63 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chrX_+_9283764 | 0.63 |
ENSMUST00000177926.1
|
1700012L04Rik
|
RIKEN cDNA 1700012L04 gene |
| chr13_+_99184733 | 0.62 |
ENSMUST00000056558.8
|
Zfp366
|
zinc finger protein 366 |
| chr15_+_102407144 | 0.62 |
ENSMUST00000169619.1
|
Sp1
|
trans-acting transcription factor 1 |
| chr13_+_48662989 | 0.62 |
ENSMUST00000021813.4
|
Barx1
|
BarH-like homeobox 1 |
| chrX_+_6873484 | 0.62 |
ENSMUST00000145302.1
|
Dgkk
|
diacylglycerol kinase kappa |
| chr7_+_43437073 | 0.61 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr11_+_69095217 | 0.61 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr11_-_70220794 | 0.61 |
ENSMUST00000159867.1
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr2_-_20943413 | 0.60 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr5_+_108065696 | 0.60 |
ENSMUST00000172045.1
|
Mtf2
|
metal response element binding transcription factor 2 |
| chr3_+_98222148 | 0.59 |
ENSMUST00000029469.4
|
Reg4
|
regenerating islet-derived family, member 4 |
| chrX_+_159840463 | 0.59 |
ENSMUST00000112451.1
ENSMUST00000112453.2 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr17_+_47630690 | 0.59 |
ENSMUST00000024779.8
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr4_-_99654983 | 0.59 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr7_+_121083414 | 0.59 |
ENSMUST00000168311.1
ENSMUST00000171880.1 |
Otoa
|
otoancorin |
| chr8_+_57511833 | 0.59 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr8_-_27202542 | 0.58 |
ENSMUST00000038174.6
|
Got1l1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr8_+_123332676 | 0.58 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr3_+_96680093 | 0.58 |
ENSMUST00000130429.1
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr2_-_91931675 | 0.58 |
ENSMUST00000111309.1
|
Mdk
|
midkine |
| chr14_+_47373813 | 0.58 |
ENSMUST00000142734.1
ENSMUST00000150290.1 ENSMUST00000144794.1 ENSMUST00000146468.1 |
Lgals3
|
lectin, galactose binding, soluble 3 |
| chr10_+_70868633 | 0.58 |
ENSMUST00000058942.5
|
4930533K18Rik
|
RIKEN cDNA 4930533K18 gene |
| chr2_-_160619971 | 0.58 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr12_+_51348370 | 0.58 |
ENSMUST00000121521.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr6_+_141249161 | 0.57 |
ENSMUST00000043259.7
|
Pde3a
|
phosphodiesterase 3A, cGMP inhibited |
| chr14_+_32165784 | 0.57 |
ENSMUST00000111994.3
ENSMUST00000168114.1 ENSMUST00000168034.1 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr16_-_20426322 | 0.57 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr14_+_62292475 | 0.57 |
ENSMUST00000166879.1
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr6_+_125067913 | 0.57 |
ENSMUST00000088292.5
|
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr11_+_4620067 | 0.57 |
ENSMUST00000109941.1
|
Gm11032
|
predicted gene 11032 |
| chr11_+_32205483 | 0.57 |
ENSMUST00000121182.1
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr11_-_97996171 | 0.57 |
ENSMUST00000042971.9
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr5_+_143548700 | 0.56 |
ENSMUST00000169329.1
ENSMUST00000067145.5 ENSMUST00000119488.1 ENSMUST00000118121.1 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr12_+_32378692 | 0.56 |
ENSMUST00000172332.2
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr7_+_28982832 | 0.56 |
ENSMUST00000085835.6
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr4_+_127169131 | 0.55 |
ENSMUST00000046659.7
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr6_-_52158292 | 0.55 |
ENSMUST00000000964.5
ENSMUST00000120363.1 |
Hoxa1
|
homeobox A1 |
| chr6_+_4504814 | 0.55 |
ENSMUST00000141483.1
|
Col1a2
|
collagen, type I, alpha 2 |
| chr6_-_82774448 | 0.55 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr18_+_34624621 | 0.55 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr3_+_145292472 | 0.55 |
ENSMUST00000029848.4
ENSMUST00000139001.1 |
Col24a1
|
collagen, type XXIV, alpha 1 |
| chr6_+_123229843 | 0.55 |
ENSMUST00000112554.2
ENSMUST00000024118.4 ENSMUST00000117130.1 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr7_+_141228766 | 0.55 |
ENSMUST00000106027.2
|
Phrf1
|
PHD and ring finger domains 1 |
| chr3_-_108085346 | 0.55 |
ENSMUST00000078912.5
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr2_-_73452666 | 0.55 |
ENSMUST00000151939.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_+_103860265 | 0.54 |
ENSMUST00000029433.7
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr1_-_33669745 | 0.54 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr6_-_83527452 | 0.54 |
ENSMUST00000141904.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr14_+_53616315 | 0.54 |
ENSMUST00000180972.1
ENSMUST00000185023.1 |
Trav12-2
|
T cell receptor alpha variable 12-2 |
| chr16_+_20696175 | 0.54 |
ENSMUST00000128273.1
|
Fam131a
|
family with sequence similarity 131, member A |
| chr6_+_35252654 | 0.54 |
ENSMUST00000152147.1
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_-_120643643 | 0.53 |
ENSMUST00000141254.1
ENSMUST00000170556.1 ENSMUST00000151876.1 ENSMUST00000026133.8 ENSMUST00000139706.1 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr12_-_4841583 | 0.53 |
ENSMUST00000020964.5
|
Fkbp1b
|
FK506 binding protein 1b |
| chr15_+_80091320 | 0.53 |
ENSMUST00000009728.6
ENSMUST00000009727.5 |
Syngr1
|
synaptogyrin 1 |
| chr1_+_87404916 | 0.53 |
ENSMUST00000173152.1
ENSMUST00000173663.1 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr15_-_76918010 | 0.53 |
ENSMUST00000048854.7
|
Zfp647
|
zinc finger protein 647 |
| chrX_+_99003224 | 0.53 |
ENSMUST00000149999.1
|
Stard8
|
START domain containing 8 |
| chr6_-_83527773 | 0.53 |
ENSMUST00000152029.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_-_132776855 | 0.52 |
ENSMUST00000106168.1
|
Fam53b
|
family with sequence similarity 53, member B |
| chr7_+_128062698 | 0.52 |
ENSMUST00000119696.1
|
Itgam
|
integrin alpha M |
| chr7_+_29309429 | 0.52 |
ENSMUST00000137848.1
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr4_-_129573637 | 0.52 |
ENSMUST00000102596.1
|
Lck
|
lymphocyte protein tyrosine kinase |
| chrX_+_139210031 | 0.52 |
ENSMUST00000113043.1
ENSMUST00000169886.1 ENSMUST00000113045.2 |
Mum1l1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr7_-_132813715 | 0.52 |
ENSMUST00000134946.1
|
Fam53b
|
family with sequence similarity 53, member B |
| chr7_+_43427622 | 0.52 |
ENSMUST00000177164.2
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr18_-_47333311 | 0.51 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr6_-_28261907 | 0.51 |
ENSMUST00000115320.1
ENSMUST00000123098.1 ENSMUST00000115321.2 ENSMUST00000155494.1 |
Zfp800
|
zinc finger protein 800 |
| chr1_+_9848375 | 0.51 |
ENSMUST00000097826.4
|
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr9_-_60522017 | 0.51 |
ENSMUST00000140824.1
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.7 | 2.0 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.6 | 0.6 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.4 | 2.2 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.4 | 1.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 1.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.4 | 2.2 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.4 | 1.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.3 | 1.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.3 | 1.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 1.0 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.3 | 2.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.3 | 0.9 | GO:2000458 | astrocyte chemotaxis(GO:0035700) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.3 | 0.9 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.3 | 0.9 | GO:0035926 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 2.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 1.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 1.6 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 0.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 | 1.6 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 0.8 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.2 | 1.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 0.8 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 1.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.6 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.6 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.2 | 2.7 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 0.5 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.2 | 0.7 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 | 1.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.2 | 0.5 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 1.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.8 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 1.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.9 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.4 | GO:0042939 | glutathione transport(GO:0034635) oligopeptide transmembrane transport(GO:0035672) tripeptide transport(GO:0042939) |
| 0.1 | 0.6 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 | 0.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.6 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 1.0 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.1 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.5 | GO:1904925 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.8 | GO:0044838 | cell quiescence(GO:0044838) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.3 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 1.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.3 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.3 | GO:0045004 | leading strand elongation(GO:0006272) DNA replication proofreading(GO:0045004) |
| 0.1 | 0.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.3 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.5 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.3 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.1 | 0.3 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.1 | 0.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.6 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.3 | GO:2000722 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.5 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 | 0.3 | GO:0010716 | regulation of collagen catabolic process(GO:0010710) negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.1 | 0.3 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.2 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 0.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.3 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.2 | GO:0036088 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.2 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.1 | 0.2 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 0.5 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.8 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.4 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 1.8 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.2 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.8 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 2.6 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.3 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.4 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.5 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 2.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.3 | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.5 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) |
| 0.0 | 0.4 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.0 | 0.4 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 2.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.5 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.4 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 3.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 | 2.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 2.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 2.8 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.5 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 1.3 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.9 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 1.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 1.3 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) strand invasion(GO:0042148) |
| 0.0 | 0.2 | GO:1904378 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.2 | GO:2000643 | negative regulation of myelination(GO:0031642) positive regulation of early endosome to late endosome transport(GO:2000643) negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.3 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.3 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 1.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.6 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.4 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.1 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.5 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 1.4 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 3.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 0.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 0.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 0.5 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 2.7 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.5 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.6 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.1 | 2.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.4 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 2.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.4 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.7 | 3.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.7 | 2.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.6 | 2.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 1.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.3 | 0.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 0.9 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.3 | 1.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 0.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 0.6 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 1.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 0.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 1.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.9 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.1 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.3 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.2 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.5 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 1.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 3.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 2.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 2.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |